or start from open source methods. Learn more about OneLab softwareUse OneLab
RapiZyme RNase Digestion for Oligo Mapping Workflows

This basic method provides the core methodology for translating a workflow into OneLab-executable script(s) as an attempt to fully or semi-automate a specific procedure. It demonstrates the benefits of automation and highlights OneLab capabilities and best practices to promote solution adoption, helping transition from manual to a more automated approach. It can be used alone or serves as a building block for a more complex workflow and is easily adaptable to users' requirements.
Overview
Long RNA molecules, such as CRISPR sgRNA and mRNA, require advanced characterization to verify their identity, sequences, and purity. Their complex functionality, structural modifications, and the presence of variants make them prime candidates for LC-MS analysis. Due to their length, these RNAs require digestion with endonucleases before analytical characterization. The resulting digestion products can then be effectively analyzed using ion-pairing reversed-phase (IPRP) or hydrophilic interaction liquid chromatography (HILIC) coupled with mass spectrometry (MS).
Waters RapiZyme MC1 and RapiZyme Cusativin provide controlled and reproducible RNA cleavage with targeted dinucleotide specificities [1]. RapiZyme MC1 efficiently cleaves at [A/U/C]pU dinucleotide bonds with minor cleavages at Cp[A/G] bonds, while RapiZyme Cusativin most efficiently cleaves at Cp[U/A/G] dinucleotide residue sequences with minor cleavages at [A/G]pU and Up[A/U] bonds (see Figure 1). The unique cleavage patterns produced by these endoribonucleases generate a wide variety of RNA digestion products with overlapping RNA products that facilitate modification mapping and improved sequence coverage. While this procedure was developed with sgRNA as the sample type, other RNAs such as messenger RNA (mRNA) may be applicable as well with protocol tuning.
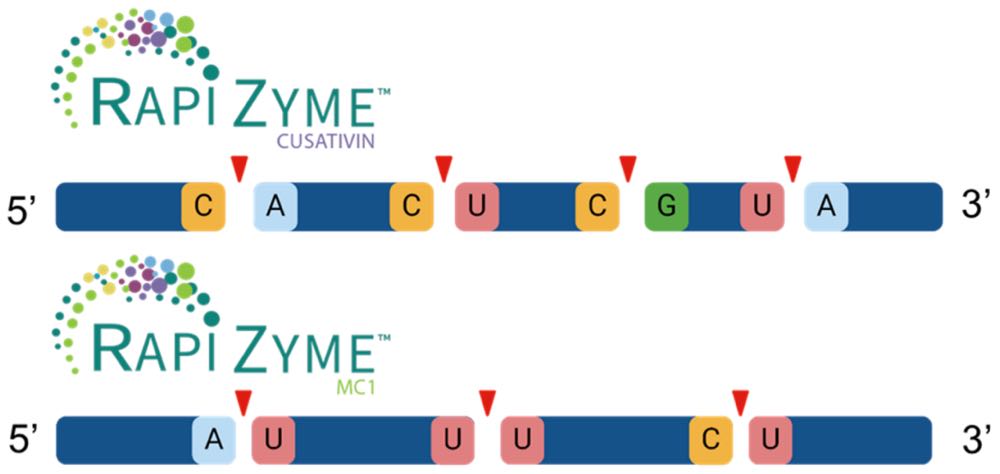
Figure 1: Example cartoon showing the primary cleavage location of top) RapiZyme Cusativin at Cp[A/U/G] and UpA and bottom) RapiZyme MC1 [A/U/C]pU dinucleotide sites. See section VI for of the RapiZyme RNases Care & Use Manual (Lit 720008488) more information on each enzyme specificities.
Prior to analysis, the RNA samples, digestion buffer, and reconstituted enzyme are combined and incubated at 30 °C to perform RNA digestion. After completion of digestion the enzyme is heat-inactivated, and the sample is transferred from a PCR tube to a 300 µL QuanRecovery 12 x 32 mm Screw Neck vial (p/n 186009186). The generic procedure for RapiZyme MC1 and Cusativin digestion is depicted in Figure 2.
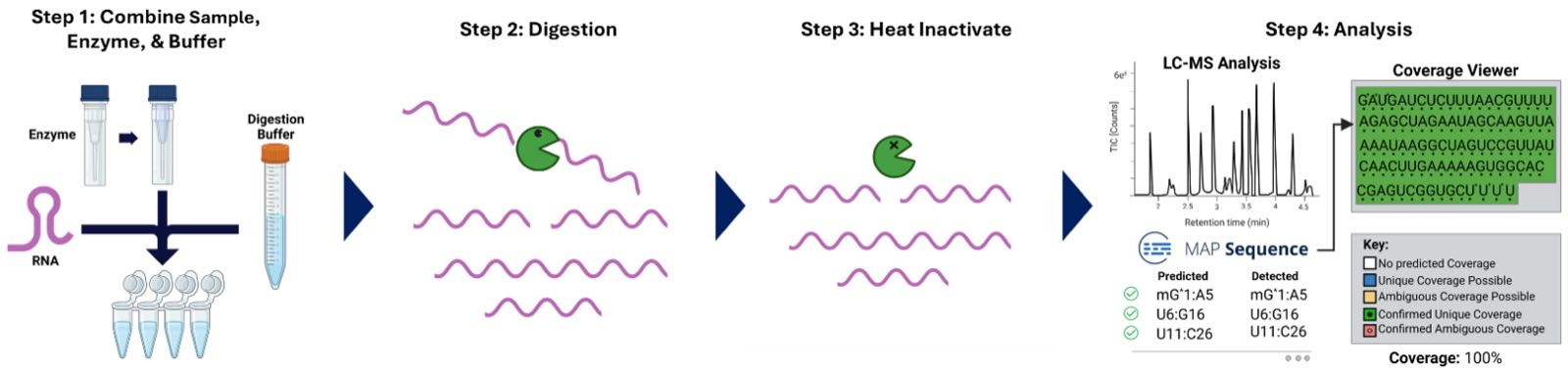
Figure 2: Schematic workflow of the RNA digestion with RapiZyme RNases (MC1 and Cusativin) and data analysis through waters_connectTM app MAP Sequence.
Assay notes
This protocol was designed for the digestion of 8 RNA samples but may be easily modified as needed. It should also be noted that the procedure was designed with the Waters sgRNA LC-MS Standard (186011357) comprised of a 100 nucleotide RNA long sequence complementary to the mouse GATA2 gene. This procedure utilizes a 10 Units to 1 µg enzyme to substrate ratio and sample incubation of 15 minutes. Longer RNA sequences may require a modified enzyme to substrate ratio or longer incubation time for more complete digestion.
The pH of the 200 mM ammonium acetate buffer recommended for RapiZyme Cusativin (pH 9) and RapiZyme MC1 (pH 8) is recommended based on a kinetic study in which various pHs were compared. Deviation from the recommended pH may impact enzymatic specificity, activity, and digestion product length. Similarly, the reaction temperature of 30 °C was found to be optimal for maximizing on-target enzyme activity.
When working with RNA, it is important to follow sterile laboratory techniques to prevent degradation. All reagents used were RNase-free certified.

Figure 3: Andrew+ OneLab deck set up for the RNA digestion protocol. Samples and reagents locations on Andrew+ deck:
(1) Tip Insertion Domino with 10-300 µL tips, (2) Tip Insertion Domino with 0.2-10 µL tips, (3) 96 PCR Domino with 8-well PCR tube strip (final position for PCR tubes with samples post centrifugation), (4) Collection Labware Rack Domino and 12 mm Vial Collection Rack with QuanRecovery 12 x 32 mm Screw Neck Vial 300 µL, (5) Conical Microtube Domino with RapiZyme MC1 or Cusativin, (6) 15 mL Centrifuge Tube Domino with 15 mL conical centrifuge tube, (7) 96-PCR Peltier+ Connected Device with 8-well PCR tube strip (starting position for PCR tubes with samples).
RapiZyme RNase automation with Andrew+ generates comparable digests to the manual workflow, as demonstrated by the TUV chromatograms of sgRNA digests prepared using automated and manual protocols (Figure 4). The LC-MS data was processed in waters_connect MAP Sequence application which automatically detect the presence or absence the RNA digestion products [2] that were generated in silico in mRNA Cleaver. Both the manual and automated MC1 and Cusativin digestions achieved 100% coverage for GATA2 sgRNA.
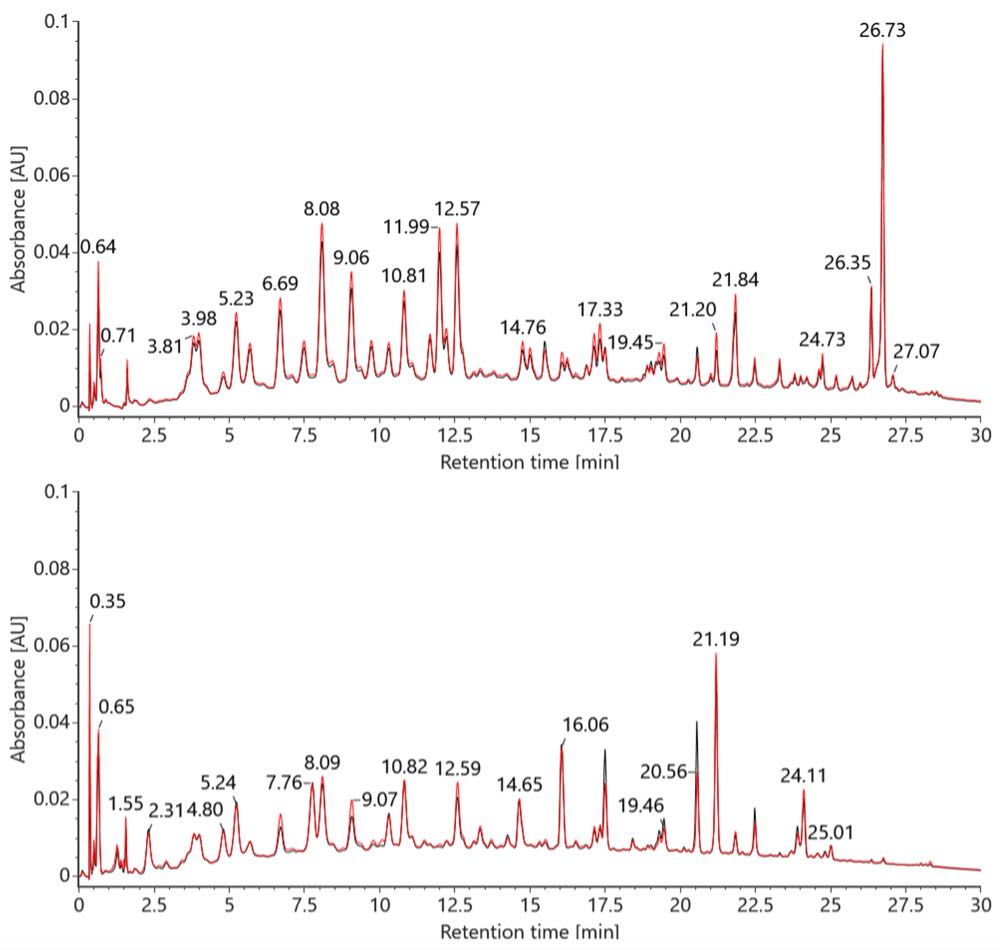
Figure 4: TUV chromatograms of GATA2 sgRNA digest prepared for MC1 (top) and Cusativin (bottom) using the manual (black) and automated (red) RapiZyme RNase procedure.
Protocols specifications
RapiZyme MC1, 8 Samples & RapiZyme Cusativin, 8 Samples:
- Estimated time of execution: 45 min
- Hands-on time: 2min (centrifuge + capping steps)
- Tip consumption:
- 8× 0.2 – 10 µL tips
- 16× 10 – 300 µL tips
Ordering information
Andrew+ System Components: Dominos, Devices, Electronic Pipettes & Tips
- Andrew+ Pipetting Robot
- OneLab software
- 2× Tip Insertion System Domino | p/n 186009612
- 96 PCR Domino | p/n 186010088
- Collection Labware Rack Domino | p/n 186010523
- 12 mm Vial Collection Rack | p/n 186010531
- Conical Microtube Domino | p/n 186009601
- 15 mL Centrifuge Tube Domino | p/n 186010087
- 96-PCR Peltier+ | p/n 186009592
- Andrew Alliance Bluetooth Electronic Pipette, 1-ch 10 μL | p/n 186009769
- Andrew Alliance Bluetooth Electronic Pipette, 1-ch 300 μL | p/n 186009606
Recommended consumables
- FrameStrip® 8-well PCR tube strip with caps (Azenta) | p/n 4ti-0786/R
- 96× FrameStrip® adapter (Azenta) | p/n 4ti-0370
- RapiZyme RNase MC1 | p/n 186011190
- RapiZyme Cusativin | p/n 186011192
- RNase-free water
- 5M ammonium acetate (Invitrogen) or equivalent | p/n AM9071
- Falcon® 15 mL centrifuge tube or equivalent
- Waters QuanRecovery™ 300 µL 12x32mm screw neck vial | p/n 186009186
References
- Addepalli, B., Johnston, T., Reidy, C., Lauber, M., Tunable Digestions of RNA Using RapiZyme™ RNases to Confirm Sequence and Map Modifications. Waters Application Note. No. 720007894, October 2024. https://www.waters.com/nextgen/en/library/application-notes/2023/automated-proteinworks-sample-preparation-using-andrew-pipetting-robot-for-gelatin-speciation-analysis-with-acquity-premier-uplc-and-xevo-tq-absolute.html.
- Doneanu, C., Preston, C., Gorton, M., Johnson, T., Addepalli, B.,Yu, Y,. RNA Digestion Product Mapping Using an Integrated UPLC-MS and Informatics Workflow. Waters Application Note. No. 720008553, October 2024. https://www.waters.com/nextgen/en/library/application-notes/2024/rna-digestion-product-mapping-using-an-integrated-uplc-ms-and-informatics-workflow.html.
Protocols
Contact info

 This is an open access protocol distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited.
This is an open access protocol distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited. 