or start from open source methods. Learn more about OneLab softwareUse OneLab
ProNex® Dual DNA Size Selection
This example method provides a freely adjustable framework for measuring the adaptability of the OneLab environment to workflows from different application fields. It helps to understand various nuances of the code-free, universal protocol designer and provides general indications as to the feasibility of a project. Scripts generally require fine adjustment to correct for variables and support specific labware implementation.
Overview
Need and Goal of Dual Size-Selective Purification
NGS library preparation requires not only the ability to clean DNA samples (removal of primers, adapters, reaction buffer, etc...) but also to selectively retain only DNA fragments of the desired size. Very large and small DNA sequences outside the desired range can interfere with the proper sequencing process. Therefore, their elimination is critical to avoid unwanted reads. Accurate size selection of your library is crucial for obtaining consistent sequencing data.
Limitations of Current Methods for Dual DNA Size Selection
Dual-size selection of DNA fragments enables the removal of DNA sequences above and below a defined size range. It is useful for whole-genome or amplicon libraries to eliminate undesired fragments or off-target amplicons, respectively. Current methods suffer from various limitations, most importantly:
- Loss of DNA during multiple purification steps. This implies the use of greater amounts of starting sample or additional PCR cycles,
- High viscosity of resin resulting in pipetting inaccuracy and consequently low reproducibility,
- Slow magnetic bead response, thereby requiring longer incubation time. Furthermore, some DNA-bound resin may remain in suspension while on the magnet and subsequently get eliminated during supernatant removal,
- Reduced specificity of size selection. In other words, current methods struggle with eliminating high molecular weight DNA that can interfere with proper sequencing.
ProNex® Dual DNA Size-Selection Protocol
The ProNex® Dual DNA Size-Selection protocol allows fast and selective magnetic resin-based purification of a population of double-stranded DNA (dsDNA) fragments centered around a desired size using proprietary ProNex® magnetic resin. Fragments above and below the desired size range are removed from the final library. The average insert size of the population ranges from 150 bp to 575 bp resulting in a target library of 275 bp to 700 bp in size, respectively. The target library size is the sum of the average insert size and a 125 bp sequence representing the average total bp length of the two NGS adapters. The size range and center point of the population can be adjusted based on the user/application needs.
The Dual DNA Size-Selection typically involves two selection steps using ProNex® chemistry (Figure 1). The desired average size can be achieved by varying the ProNex® chemistry:sample (v/v) ratio. The higher the ratio required to achieve a desired size, the smaller the purified dsDNA fragments, and inversely. It is worth noting that the size selection outcome can vary depending on the type of starting samples. Therefore, accurate and reproducible size selection may require optimization before attempting to purify a crucial sample.
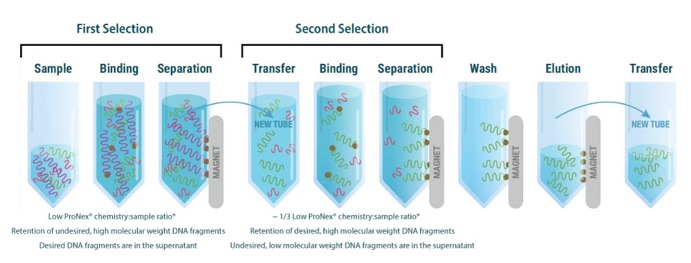
Figure 1: A schematic illustration of the procedure for size-selective purification of dsDNA fragments centered around a desired size.
The ProNex® Dual DNA Size-Selection not only efficiently removes off-target fragments (low and high molecular weight DNA) but also enriches the library with target fragment sizes. This protocol offers several benefits:
- Enhanced specificity of size selection with more complete removal of undesired DNA from the final library,
- High recovery of input DNA resulting in increased final library yield,
- Low viscosity of resin ensuring accurate pipetting, better reproducibility and compatibility with automation,
- Fast magnetic bead response time.
Protocols
Contact info

 This is an open access protocol distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited.
This is an open access protocol distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited. 