or start from open source methods. Learn more about OneLab softwareUse OneLab
Plasmid Purification and Clean-up

This basic method provides the core methodology for translating a workflow into OneLab-executable script(s) as an attempt to fully or semi-automate a specific procedure. It demonstrates the benefits of automation and highlights OneLab capabilities and best practices to promote solution adoption, helping transition from manual to a more automated approach. It can be used alone or serves as a building block for a more complex workflow and is easily adaptable to users' requirements.
Overview
NucleoSpin Plasmid ─ Plasmid DNA Extraction (Miniprep)
The NucleoSpin Plasmid miniprep kit is designed for the rapid (25 minutes), high-yield (up to 45 μg), small-scale preparation of high-purity plasmid DNA. The NucleoSpin Plasmid miniprep columns contain a unique silica membrane that binds up to 50 μg of plasmid DNA. The purified DNA is suitable for transfection, cloning, sequencing, PCR, transformation, and restriction analysis.
► Plasmid DNA Purification Using NucleoSpin Plasmid
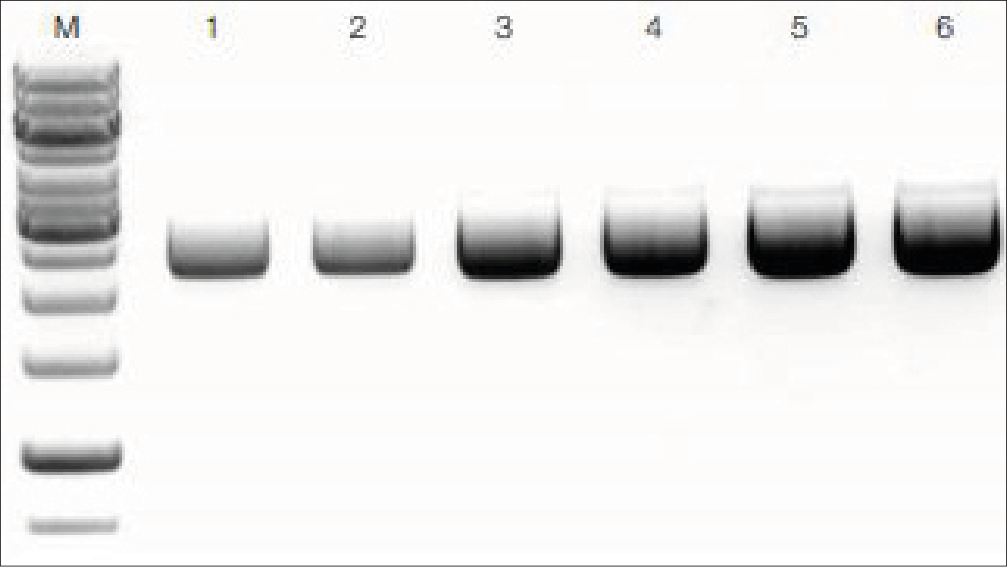
Figure 1: Plasmid DNA isolation (pUC18) from E. coli DH5α competent cells using NucleoSpin Plasmid. For plasmid DNA isolation, 2 mL (lane 1–2), 5 mL (lane 3–4), and 8 mL (lane 5–6) volumes of LB cultures were used and analyzed on an agarose gel (2 μL of each eluate). Lane M: marker or DNA ladder.
► NucleoSpin Plasmid Miniprep Workflow
Harvested bacteria are resuspended and processed by SDS/alkaline lysis. A high-salt buffer is added to neutralize the lysate and to create appropriate conditions for DNA binding to the silica membrane. After centrifugation, the clear supernatant is loaded onto a NucleoSpin Plasmid miniprep spin column. Contaminants like salts and macromolecular cellular components are removed by simple washing with ethanolic Buffer A4. Additional washing with Buffer AW is recommended for the following demands:
- Complete removal of high levels of endonucleases (e.g., from wild-type E. coli strains), with Buffer AW prewarmed to 50°C,
- Processing 5–10 mL of bacterial culture,
- Long read length in DNA sequencing,
- Better performance of critical enzymatic reactions.
Highly pure plasmid miniprep DNA is finally eluted in 50 μL of slightly alkaline Buffer AE (5 mM Tris/HCl, pH 8.5).
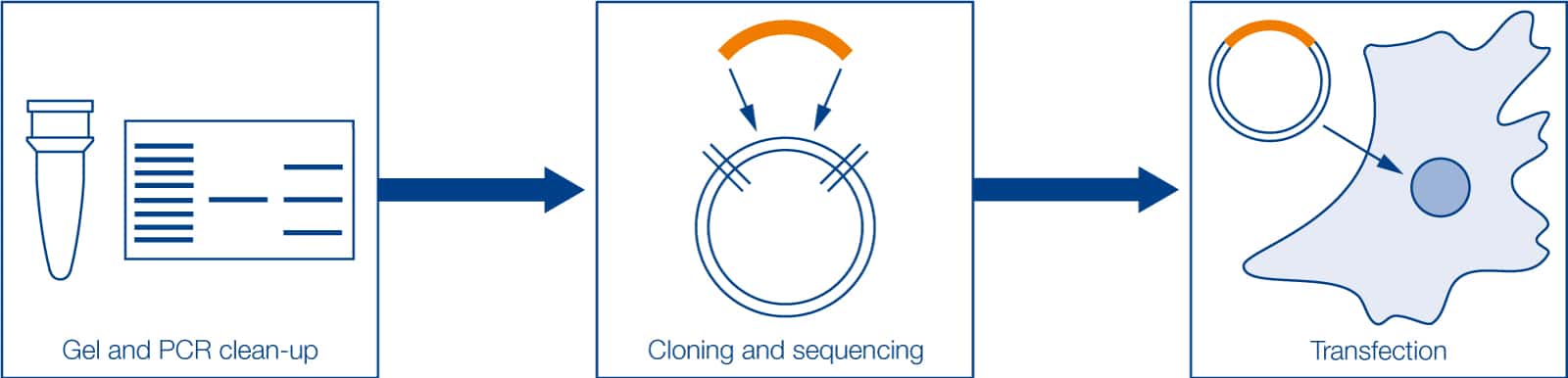
Figure 2: Cloning and transfection workflow. Cloning methods require several clean-up steps to recover digested fragments from the agarose gel, i.e., DNA/PCR insert and DNA vector, in order to reconstitute the plasmid with the target gene. Following transformation, bacteria carrying the recombinant plasmid are selected and grown up in a rich medium. As bacteria reproduce, they replicate the plasmid, generating millions of copies of the recombinant DNA molecule. Bacteria are harvested and plasmids are extracted and purified. Finally, after verification of the recombinant plasmid construction (by colony PCR, restriction digestion, and/or sequencing), plasmids are ready to be used in cellular biology research, for instance, cell transfection and expression of the target gene.
NucleoSpin Gel and PCR Clean-up ─ PCR Purification and Gel Extraction
With the NucleoSpin Gel and PCR Clean-up kits, you get two applications in one. The supplied columns, which are suitable for gel extraction as well as PCR purification, bind DNA to a silica membrane in the presence of a chaotropic salt. Once the binding mixture is loaded onto the column, contaminants are removed by simple wash steps with ethanolic Wash Buffer NT3, providing rapid DNA purification. You can isolate fragments from 50 bp–20 kb in minimal elution volumes with recovery rates of 70–95%. Downstream applications include sequencing, ligation, labeling, cloning, and restriction enzyme digestion.
► Binding Buffer with a pH Indicator
The optimal pH to bind even small DNA fragments to the silica membrane is about 5–6. Binding Buffer NTI is sufficiently buffered to maintain this pH. However, to be sure that the pH is right even for samples with extreme alkaline pH or high buffer concentration, a pH indicator has been added (Figure 3). The pH indicator does not interfere with DNA binding and is completely removed during the purification. The yellow color is also beneficial for gel extractions, making it easy to identify undissolved pieces of agarose. To restore the correct conditions, add more Buffer NTI or 4 M sodium acetate (NaAc, pH 5.0), or small amounts of hydrochloric acid (HCl) until the color switches back to yellow.

Figure 3: The range of colors displayed by the pH indicator in Binding Buffer NTI.
► High Recovery Rates for PCR Clean-up
The NucleoSpin Gel and PCR Clean-up kit showed a high recovery rate for a wide range of fragment sizes, down to 50 bp (Figure 4). DNA fragments of different sizes were purified from a standard PCR buffer using the kit, with elution volumes of 15 μL, 30 μL, and 50 μL. DNA was analyzed and quantified using an Agilent Bioanalyzer 2100.
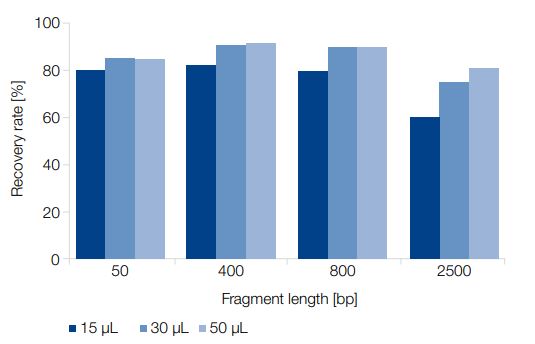
Figure 4: High recovery rates for PCR clean-up of fragments down to 50 bp.
Ordering Information ─ Kit Consumables by MACHEREY-NAGEL
- NucleoSpin Plasmid - Mini spin kit for purifying plasmid DNA I REF: 740588.10/.50/.250
- NucleoSpin Gel and PCR Clean-up - Mini spin kit for gel extraction and PCR clean-up I REF: 740609.10/.50/.250
Find more information about MACHEREY-NAGEL plasmid purification kits
Tips and Tricks to optimize your PCR clean-up and gel extraction
Protocols



Contact info

 This is an open access protocol distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited.
This is an open access protocol distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited. 