or start from open source methods. Learn more about OneLab softwareUse OneLab
Phenol-Chloroform gDNA Extraction

This example method provides a freely adjustable framework for measuring the adaptability of the OneLab environment to workflows from different application fields. It helps to understand various nuances of the code-free, universal protocol designer and provides general indications as to the feasibility of a project. Scripts generally require fine adjustment to correct for variables and support specific labware implementation.
Overview
The most basic of all procedures in molecular biology is the extraction of genomic DNA, typically using the phenol-chloroform method (Figure 1). High-quality genomic DNA is used in many applications, such as cloning, Southern blot, PCR and sequencing library preparation. Given the prevalence of home-brew solutions, the ability to develop and share a standardized protocol, as can be achieved with OneLab, is especially valuable.
The protocol described here is intended to isolate genomic DNA from bacterial cells. Cells are initially grown in an appropriate culture medium at an optimal temperature. For maximum yield, cells are harvested at the late-log phase by centrifugation and resuspended in TE buffer (Tris:EDTA, pH 8.0) and SDS (Sodium dodecyl sulfate). Then, cells are disrupted mechanically by passing them through a syringe needle. SDS, a detergent surfactant, is used to remove membrane lipids, while the TE buffer prevents DNases from degrading DNA through chelating divalent cations, such as Mg2+ (superscript). Furthermore, proteinase K and RNase A are added to the mixture for the enzymatic digestion of proteins and RNA molecules, respectively. The phenol:chloroform solution is used to denature and separate proteins from DNA. It promotes the partitioning of lipids and other cellular debris into the organic phase, leaving DNA in the aqueous upper phase (Figure 1). The denatured proteins form a middle layer between the aqueous and organic phases. Following centrifugation, the aqueous phase containing the DNA is transferred to a new tube in order to carry out DNA precipitation using isopropanol. Upon addition of isopropanol, nucleic acid molecules aggregate, and a white precipitate is formed. Additionally, sodium acetate is used to improve the precipitation of DNA by increasing ionic strength, thus rendering the DNA less soluble. The DNA pellet is finally washed in 70% ice-cold ethanol to remove residual salt, then air-dried and resuspended in DNase-free water.
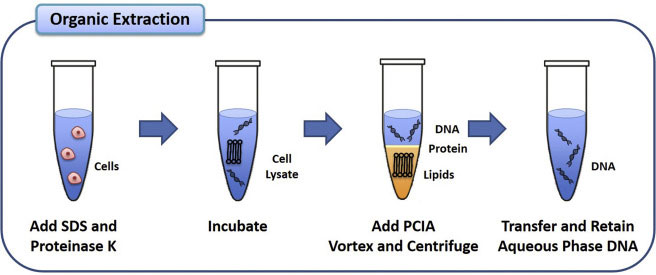
Figure 1: Schematic illustration showing the four major steps of the DNA extraction procedure using phenol:chloroform.
The addition of phenol:chloroform to the cell lysate partitions DNA to the aqueous phase, while lipids and denatured proteins are partitioned to the organic phase and the middle layer, respectively.
This extraction technique generally yields high-purity DNA. PCIA, phenol:chloroform:isoamyl alcohol.
Image adapted from McKiernan, H. E. & Danielson, P. B. Molecular Diagnostics (Third Edition), 371–394. Academic Press, 2017.
Discover another variant of the phenol-chloroform extraction protocol
Protocols
Contact info

 This is an open access protocol distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited.
This is an open access protocol distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited. 