or start from open source methods. Learn more about OneLab softwareUse OneLab
PeptideWorks Tryptic Protein Digestion Sample Prep
This advanced method offers an end-to-end solution developed around an advanced application, a specialized kit, or an analytical system. It delivers complete functional, ready-to-use protocols that are qualitatively and quantitatively assessed for consistency, executability, and repeatability, usually without needing to make any changes. It operates per batch size and is scalable to accommodate changing requirements.
Overview
Reliable Sample Preparation with Waters Automated PeptideWorks™ Protocol
Peptide mapping is valuable in biopharmaceutical research and development because it delivers comprehensive information about the primary structure of a protein (1). Peptide mapping is used in QC and bioprocess development for batch release, stability testing, and clone selection. Generally, the peptide map of a test article is compared to that of a reference material; sample-to-sample reproducibility is therefore imperative for the detection of real changes between the test and reference materials. Reliable sample preparation is the most critical factor in generating meaningful peptide mapping data.
Sample preparation for peptide mapping is one of the most complex processes for the routine analysis of therapeutic proteins. Prior to analysis, samples are treated in an alkaline buffer at high temperatures and enzymatically digested to generate peptides. These conditions can result in method-induced peptide modifications, over- or under-digestion of the protein, and autolysis of the proteolytic enzyme, each of which complicate data analysis and interpretation. Moreover, protein digestion is often plagued with long digestion times, potentially slowing down critical drug development and release work.
Waters PeptideWorks kit delivers fast and reliable sample preparation for routine peptide mapping of therapeutic proteins. The sample preparation kit is centred around RapiZyme™ trypsin, Waters’ homogeneously methylated, recombinant porcine trypsin. RapiZyme trypsin enables uncompromising speed and digestion cleanliness through its inordinately high autolysis resistance, high purity, and improved activity (2, 3, 4). By virtually eliminating autolysis, RapiZyme trypsin can be used at high concentrations to achieve faster digestion without requiring high temperatures or sacrificing digestion completeness. The reagents and reaction conditions used in the PeptideWorks kit were optimized to enable fast and complete digestion of proteins by RapiZyme trypsin.
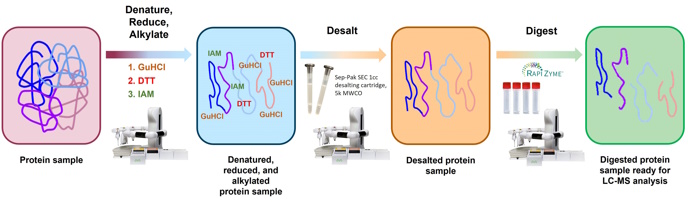
Figure 1: Workflow for the automated preparation of digested peptides using the PeptideWorks sample preparation kit.
Automated PeptideWorks Sample Preparation Procedure
The automated PeptideWorks workflow is illustrated in Figure 1. The automated workflow is split into two protocols: protocol A executes denaturation, reduction, and alkylation and protocol B executes digestion. In protocol A, the Andrew+ pipetting robot is used to denature the protein with guanidine hydrochloride (GuHCl), reduce it with dithiothreitol (DTT), and alkylate it with iodoacetamide (IAM). Extraction+ is then used with Waters Sep-Pak® SEC desalting cartridges to remove GuHCl, DTT, and IAM from the reaction. In protocol B, the protein is digested at 37°C with RapiZyme trypsin and the reaction is quenched with formic acid. If including a concentration normalization step of the desalted protein, the concentration can be measured offline, imported into OneLab software, and normalized in protocol B using the Andrew+ Pipetting Robot.
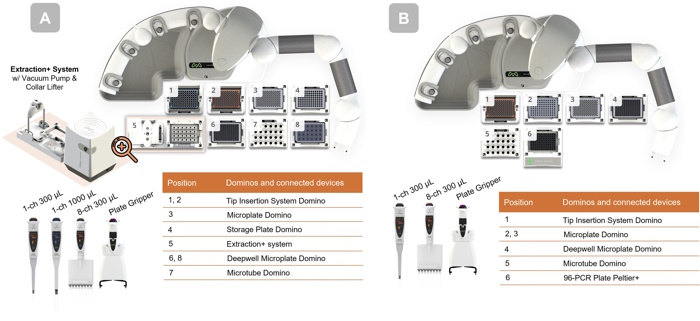
Figure 2: Andrew+ deck layouts for the 24-sample automated PeptideWorks procedure. (A) Andrew+ configuration for the PeptideWorks protocol A, 24 samples, with concentration normalization. (B) Andrew+ configuration for the PeptideWorks protocol B, 24 samples, with concentration normalization.
The automated PeptideWorks workflow can accommodate up to 24 samples. The deck layouts for a full 24-sample protocol are shown in Figure 2. The total execution time for 24 samples is 2h 40min with concentration normalization and 2h 30min without concentration normalization.
PeptideWorks automation with Andrew+ generates comparable digests to the manual workflow, as demonstrated by the BPI chromatograms of NISTmAb digests prepared using automated and manual PeptideWorks protocols (Figure 3). Both protocols yield digests with high sequence coverage (>88% expected peptides), low abundance of miscleavages (<4%), and low abundance of nonspecific cleavages (<1%) (5).
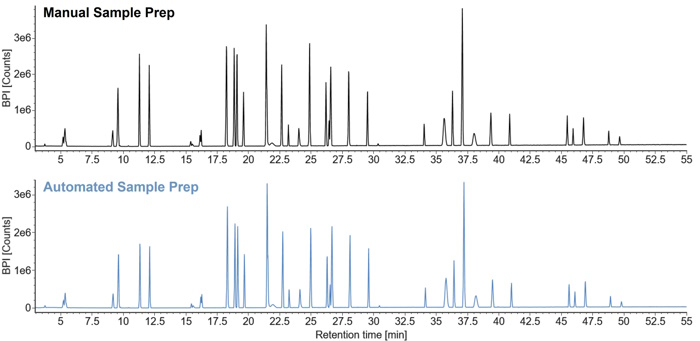
Figure 3: BPI chromatograms of NISTmAb digests prepared using the manual and automated PeptideWorks procedures.
Considerations
1- General Protocol
- This procedure was designed as a general protocol and may need to be adjusted to suit individual sample needs.
2- Amount of Digested Protein
- The digestion protocol was optimized to digest 60 pmol of protein. Significant deviations from this amount may reduce digestion efficiency and require optimization of protein concentration, RapiZyme trypsin concentration, or digestion time.
3- Additional notes
- For protocol A, the concentration normalization is recommended to ensure accurate enzyme: protein ratios during digestion
- For protocols using 4 samples, please store any unused GuHCl at 4 deg C for further use.
Protocol Specifications
▶ PeptideWorks Protocol A, 24 samples, with Conc. Norm.
- Estimated time of execution | 1h 38min 8sec
- Tip consumption | 96x 5-350 μL tips and 8x 10-1000 μL tips
▶ PeptideWorks Protocol A, 24 samples, without Conc. Norm.
- Estimated time of execution | 1h 38min 51sec
- Tip consumption | 96x 5-350 μL tips and 8x 10-1000 μL tips
▶ PeptideWorks Protocol A, 4 samples, with Conc. Norm.
- Estimated time of execution | 1h 24min 21sec
- Tip consumption | 20x 5-350 μL tips and 5x 10-1000 μL tips
▶ PeptideWorks Protocol A, 4 samples, without Conc. Norm.
- Estimated time of execution | 1h 24min 33sec
- Tip consumption | 20x 5-350 μL tips and 5x 10-1000 μL tips
▶ PeptideWorks Protocol B, 24 samples, with Conc. Norm.
- Estimated time of execution | 48min 41sec
- Tip consumption | 74x 5-350 μL tips
▶ PeptideWorks Protocol B, 24 samples, without Conc. Norm.
- Estimated time of execution | 39min 9sec
- Tip consumption | 73x 5-350 μL tips
▶ PeptideWorks Protocol B, 4 samples, with Conc. Norm.
- Estimated time of execution | 38min 47sec
- Tip consumption | 13x 5-350 μL tips
▶ PeptideWorks Protocol B, 4 samples, without Conc. Norm.
- Estimated time of execution | 38min 10sec
- Tip consumption | 12x 5-350 μL tips
ORDERING INFORMATION
Andrew+ System Components: Dominos, Devices, Electronic Pipettes & Tips
➤ PeptideWorks Protocol A, 24 Samples, with OR without Conc. Norm. – Andrew+
- 1x Microplate Domino | p/n 186009600
- 1x Storage Plate Domino | p/n 186009596
- 1x Microtube Domino | p/n 186009601
- 2x Deepwell Microplate Domino | p/n 186009597
- 2x Tip Insertion System Domino | p/n 186009612
- Extraction+ Base Kit with Plate Gripper | p/n 176005201
- Extraction+ 1 cc Cartridge Kit | p/n 176005204
- Andrew Alliance Bluetooth Electronic Pipette, 1-ch 300 μL | p/n 186009606
- Andrew Alliance Bluetooth Electronic Pipette, 1-ch 1000 μL | p/n 186009766
- Andrew Alliance Bluetooth Electronic Pipette, 8-ch 300 μL | p/n 186009607
- Sartorius, Optifit Tips, 5-350 μL (x96) | p/n 700013297
- Sartorius, Optifit Tips, 10-1000 μL (x8) | p/n 700013298
➤ PeptideWorks Protocol A, 4 Samples, with OR without Conc. Norm. – Andrew+
- 1x Microplate Domino | p/n 186009600
- 1x Storage Plate Domino | p/n 186009596
- 1x Microtube Domino | p/n 186009601
- 1x Deepwell Microplate Domino | p/n 186009597
- 1x 50mL Conical Centrifuge Tube Domino | p/n 186009614
- 2x Tip Insertion System Domino | p/n 186009612
- Extraction+ Base Kit with Plate Gripper | p/n 176005201
- Extraction+ 1 cc Cartridge Kit | p/n 176005204
- Andrew Alliance Bluetooth Electronic Pipette, 1-ch 300 μL | p/n 186009606
- Andrew Alliance Bluetooth Electronic Pipette, 1-ch 1000 μL | p/n 186009766
- Sartorius, Optifit Tips, 5-350 μL (x96) | p/n 700013297
- Sartorius, Optifit Tips, 10-1000 μL (x8) | p/n 700013298
➤ PeptideWorks Protocol B, 24 Samples, with Conc. Norm. – Andrew+
- 2x Microplate Domino | p/n 186009600
- 1x Microtube Domino | p/n 186009601
- 1x Deepwell Microplate Domino | p/n 186009597
- 1x Tip Insertion System Domino | p/n 186009612
- 96-PCR Plate Peltier+ | p/n 176004584
- Andrew Alliance Bluetooth Microplate Gripper | p/n 186009776
- Andrew Alliance Bluetooth Electronic Pipette, 1-ch 300 μL | p/n 186009606
- Andrew Alliance Bluetooth Electronic Pipette, 8-ch 300 μL | p/n 186009607
- Sartorius, Optifit Tips, 5-350 μL (x74) | p/n 700013297
➤ PeptideWorks Protocol B, 24 Samples, without Conc. Norm. – Andrew+
- 1x Microplate Domino | p/n 186009600
- 1x Microtube Domino | p/n 186009601
- 1x Deepwell Microplate Domino | p/n 186009597
- 1x Tip Insertion System Domino | p/n 186009612
- 96-PCR Plate Peltier+ | p/n 176004584
- Andrew Alliance Bluetooth Microplate Gripper | p/n 186009776
- Andrew Alliance Bluetooth Electronic Pipette, 1-ch 300 μL | p/n 186009606
- Andrew Alliance Bluetooth Electronic Pipette, 8-ch 300 μL | p/n 186009607
- Sartorius, Optifit Tips, 5-350 μL (x73) | p/n 700013297
➤ PeptideWorks Protocol B, 4 Samples, with Conc. Norm. – Andrew+
- 2x Microplate Domino | p/n 186009600
- 1x Microtube Domino | p/n 186009601
- 1x 50mL Conical Centrifuge Tube Domino | p/n 186009614
- 1x Tip Insertion System Domino | p/n 186009612
- 96-PCR Plate Peltier+ | p/n 176004584
- Andrew Alliance Bluetooth Microplate Gripper | p/n 186009776
- Andrew Alliance Bluetooth Electronic Pipette, 1-ch 300 μL | p/n 186009606
- Sartorius, Optifit Tips, 5-350 μL (x13) | p/n 700013297
➤ PeptideWorks Protocol B, 4 Samples, without Conc. Norm. – Andrew+
- 1x Microplate Domino | p/n 186009600
- 1x Microtube Domino | p/n 186009601
- 1x 50mL Conical Centrifuge Tube Domino | p/n 186009614
- 1x Tip Insertion System Domino | p/n 186009612
- 96-PCR Plate Peltier+ | p/n 176004584
- Andrew Alliance Bluetooth Microplate Gripper | p/n 186009776
- Andrew Alliance Bluetooth Electronic Pipette, 1-ch 300 μL | p/n 186009606
- Sartorius, Optifit Tips, 5-350 μL (x12) | p/n 700013297
Application Kits
1- PeptideWorks Tryptic Protein Digestion Start-Up without unfolding | p/n 176005308
- Sep-Pak SEC Desalting Cartridge 1cc Start-Up Kit | p/n 186010126
- RapiZyme Trypsin | p/n 725000692
- 1M CaCl2 Salt Concentrate, bulk | p/n 150001895
2- PeptideWorks Tryptic Protein Digestion Refill Kit without unfolding | p/n 176005309
- Sep-Pak SEC Desalting Cartridge 1cc Start-Up Kit | p/n 186010126
- RapiZyme Trypsin | p/n 725000692
- 1M CaCl2 Salt Concentrate, bulk | p/n 150001895
3- PeptideWorks Tryptic Start-Up & Column Kit | p/n 176005310
- GuHCl 5.7g bulk | p/n 150001891
- DTT 16 mg, bulk | p/n 150001889
- IAM 53 mg bulk | p/n 150001890
- Sep-Pak SEC Desalting Cartridge 1cc Start-Up Kit | p/n 186010126
- RapiZyme Trypsin | p/n 725000692
- 1M CaCl2 Salt Concentrate, bulk | p/n 150001895
- ACQUITY Premier Peptide CSH C18 Column | p/n 186009489
4- PeptideWorks Tryptic Protein Digestion Start-Up Kit | p/n 176005311
- GuHCl 5.7g bulk | p/n 150001891
- DTT 16 mg, bulk | p/n 150001889
- IAM 53 mg bulk | p/n 150001890
- Sep-Pak SEC Desalting Cartridge 1cc Start-Up Kit | p/n 186010126
- RapiZyme Trypsin | p/n 725000692
- 1M CaCl2 Salt Concentrate, bulk | p/n 150001895
5- PeptideWorks Tryptic Protein Digestion Refill Kit | p/n 176005312
- GuHCl 5.7g bulk | p/n 150001891
- DTT 16 mg, bulk | p/n 150001889
- IAM 53 mg bulk | p/n 150001890
- Sep-Pak SEC Desalting Cartridges 1cc | p/n 186010127
- RapiZyme Trypsin | p/n 725000692
- 1M CaCl2 Salt Concentrate, bulk | p/n 150001895
Reagents & Chemicals (Not Provided in PeptideWorks kits)
- 18.2 MΩ water
- 1 M Tris HCl stock (to be diluted to 100 mM)
- Formic acid (to make a 1% v/v solution)
Recommended Consumables
- Axygen® 10 mL 24-square deep well V-bottom plate | p/n P-DW-10ML-24-C-S
- Axygen® 12-well reservoir, 12-channel trough | p/n RES-MW12-HP
- Falcon® 50 mL conical centrifuge tube | p/n 352070
- Fisherbrand™ Premium 1.5 mL microtube | p/n 11926955
- Grenier 96-well flat-bottom microplate | p/n 655101
- Eppendorf twin.tec® 96-well skirted LoBind® PCR plate | p/n 0030129555
- Waters QuanRecovery™ 700 μL 96-well plate | p/n 186009185
(1) Hada, V., Bagdi, A., Bihari, Z., Timari, S. B., Fizil, A., Szantay Jr. C. Recent advancements, challenges, and practical considerations in the mass spectrometry-based analytics of protein biotherapeutics: A viewpoint from the biosimilar industry. Jour. of Pharm. and Biom. Anal. 2018, 161, 214-2382.
(2) Ippoliti, S., Zampa, N., Yu, Y. Q., Lauber, M. A. Versatile and Rapid Digestion Protocols for Biopharmaceutical Characterization Using RapiZymeTM Trypsin. (720007840EN) Waters Corporation January 2023.
(3) Finny, A. S., Zampa, N., Addepalli, B., Lauber, M. A. Fast and Robust LC-UV-MS Based Peptide Mapping Using RapiZymeTM Trypsin and IonHanceTM DFA. (720007864EN) Waters Corporation February 2023.
(4) Yang, H., Koza, S. M., Yu, Y. Q. Automated High-Throughput LC-MS Focused Peptide Mapping of Monoclonal Antibodies in Microbioreactor Samples. (720007885EN) Waters Corporation April 2023.
(5) Hanna, C. M., Danaceau, J. P., Koza, S. M., Shiner, S., Trudeau, M. Automated PeptideWorksTM Peptide Mapping using the Andrew+ Pipetting RobotTM. Waters Corporation, in development.
▌A detailed Application Note is currently under development to provide more information about the automation of PeptideWorks™ peptide map sample preparation using the Andrew+ Pipetting Robot.
Protocols

Contact info

 This is an open access protocol distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited.
This is an open access protocol distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited. 