or start from open source methods. Learn more about OneLab softwareUse OneLab
O-glycopeptide Profile Analysis

This advanced method offers an end-to-end solution developed around an advanced application, a specialized kit, or an analytical system. It delivers complete functional, ready-to-use protocols that are qualitatively and quantitatively assessed for consistency, executability, and repeatability, usually without needing to make any changes. It operates per batch size and is scalable to accommodate changing requirements.
Overview
Reducing the Complexity of O-glycosylation Analysis using the O-protease OpeRATOR
The development and manufacturing of therapeutic proteins require the analysis of many critical quality attributes (CQAs). One such potential CQA is protein glycosylation, which in many cases has proven important for the protein structure and function. However, O-glycans, as opposed to N-glycans, can be quite challenging to analyze. At the same time, there is a strong demand from the biopharma industry for robust and high-throughput analysis methods for these CQAs.
O-glycans often cluster into heavily glycosylated regions rich in serine (Ser) and threonine (Thr) residues. These regions are usually not adequately digested by common proteases such as trypsin, which makes peptide mapping approaches to characterize O-glycosylation difficult. The inherent heterogeneity of heavily O-glycosylated proteins makes analysis by mass spectrometry at the intact or subunit level challenging and requires high-resolution instrumentation. Released glycan analysis, which is commonly used to characterize N-linked glycosylation, is hampered by the lack of enzymatic tools to release intact O-glycans and has to rely on cumbersome chemical release methods. Furthermore, similar to LC-MS analysis at the intact level, these approaches do not provide site-specific information. Recently, a novel protease was identified with specificity for N-terminal digestion at Ser/Thr residues with a core 1 O-glycan. The protease, which has been commercialized under the name OpeRATOR®, generates peptides with only one or a few O-glycans, enabling detailed site-specific analysis of heavily O-glycosylated proteins by LC-MS. The relative intensities of the resulting O-glycosylated peptides can be used as a fingerprint to compare the O-glycan composition of different protein batches in an automated fashion.
Here we present an automated workflow for monitoring the O-glycopeptide profiles of heavily O-glycosylated proteins. The protocol was optimized for the TNFR fusion protein Etanercept and is based on digestion with the O-protease OpeRATOR, followed by reduction and alkylation of the disulfide bridges of the resulting peptides. The sample preparation protocol was automated on the Andrew+ Pipetting Robot and yielded peptide samples ready for analysis by LC-MS (Figure 1).
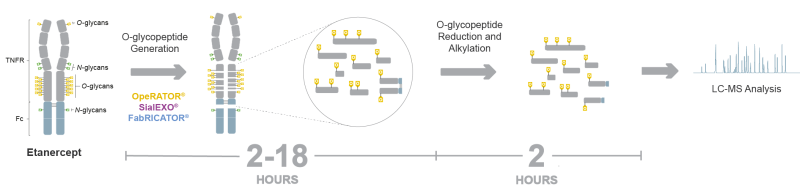
Figure 1: Automated workflow for monitoring the O-glycopeptide profile of Etanercept using the protease OpeRATOR on the Andrew+ Pipetting Robot for subsequent analysis by LC-MS.
Experimental Procedure
OpeRATOR digests N-terminally to Ser and Thr residues carrying a core 1 O-glycan and requires the presence of the sialidase SialEXO® to achieve the optimal activity. The digestion takes from 2 hours to around 18 hours, depending on the target protein. This assay was optimized for the biotherapeutic Etanercept but is applicable to many other heavily O-glycosylated proteins. Etanercept is a fusion protein where the extracellular domain of the TNFα receptor is linked to the Fc portion of human IgG1. The protein contains a cluster of up to 13 O-glycosylated Ser/Thr residues just above the hinge, making it a very difficult biopharmaceutical to characterize.
In the first protocol of this workflow, an enzyme master mix is added to the target protein samples using the Andrew+ Pipetting robot to generate the O-glycopeptides (Figure 2A). Two protocols are provided for this purpose, one protocol for Fc-Fusion proteins that uses the enzymes OpeRATOR, SialEXO, and FabRICATOR®, and the second for other proteins in which FabRICATOR is not included in the enzyme master mix. SialEXO will remove all sialic acids from the O-glycans to ensure optimal digestion by OpeRATOR, which will digest the protein backbone in the heavily O-glycosylated region of the substrate protein into small O-glycopeptides. Regions without O-glycans, such as the Fc fragment and a large N-terminal region of the TNFR domain in Etanercept, are left intact and do not show up in the subsequent analysis of the peptides. The FabRICATOR protease digests the fusion protein at one single digestion site below the hinge and thereby separates the O-glycosylated TNFR region from the Fc region to enable an analysis of the most C-terminal O-glycosylation sites. Enzymatic digestion with both protocols is performed overnight at 37°C in an incubator outside the robot working deck to avoid evaporation, leaving the robot available for other tasks during the incubation time. In the second part of the workflow, the digested protein sample is placed back into the Andrew+ Pipetting Robot for reduction and alkylation of the cysteine disulfide linkages of the peptides (Figure 2B). The yielded O-glycosylated peptides are collected in a microplate suitable for direct transfer into the autosampler of an LC-MS system.
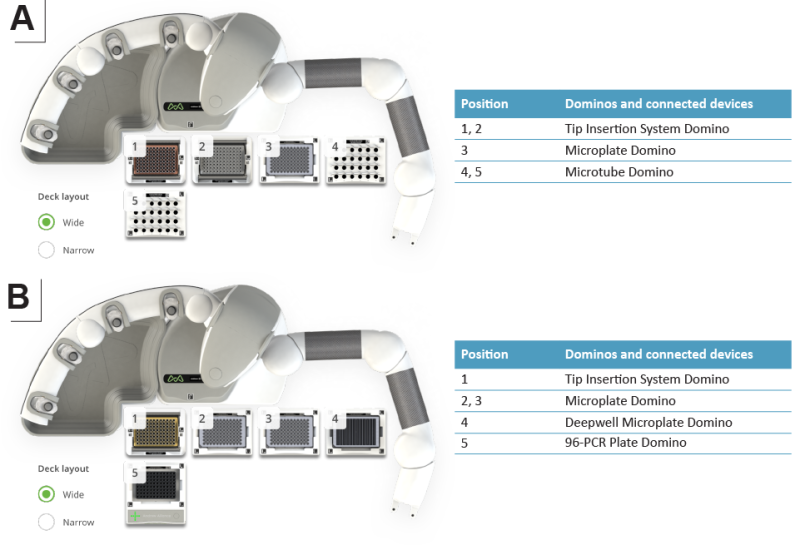
Figure 2: Domino/device positions on the Andrew+ working deck for (A) the “O-glycopeptide generation” protocol and (B) the “O-glycopeptide reduction and alkylation" protocol.
Considerations
1- The workflow has been tested on the Etanercept originator in both the formulation buffer and in 20 mM Tris buffer, pH 7.5. Please note that the formulation buffer may affect the enzymes’ efficacies and that O-glycoprofile comparisons should only be performed on proteins in similar buffers.
2- The presence of FabRICATOR in the enzyme master mix is only applicable for Fc-fusion proteins and serves to separate the Fc region from the O-glycosylated region fused to it.
3- Protocol 2 is only applicable if the target protein contains cysteine residues.
4- The workflow may need to be optimized if applied to other proteins than Etanercept.
5- Protocol 2 uses “dummy” samples in rows 7-8 to make use of the multichannel pipettes. This makes the protocol faster but leads to somewhat higher consumption of pipette tips. If this is a concern, the protocol can be adapted for use of a single-channel pipette.
6- The 350 μL 96-round well collection plate can be placed directly into the autosampler of a Waters LC-MS system. For this purpose, the plate was sealed using a polypropylene cap mat to avoid contamination and evaporation.
ORDERING INFORMATION
Andrew+ System Components: Dominos, Devices, Electronic Pipettes & Tips
➤ O-glycopeptide Generation, Fc-fusion Proteins, 2 Samples, 3 Replicates (Protocol 1/2)
- 2x Tip Insertion System Domino | p/n 186009612
- Microplate Domino | p/n 186009600
- 2x Microtube Domino | p/n 186009601
- Andrew Alliance Bluetooth Electronic Pipette, 1-ch 300 μL | p/n 186009606
- Andrew Alliance Bluetooth Electronic Pipette,1-ch 10 μL | p/n 186009769
- Sartorius, Optifit Tips, 0.1-10 μL (x6) | p/n 700013293
- Sartorius, Optifit Tips, 5-350 μL (x5) | p/n 700013297
➤ O-glycopeptide Generation, Other Proteins, 2 Samples, 3 Replicates (Protocol 1/2)
- 2x Tip Insertion System Domino | p/n 186009612
- Microplate Domino | p/n 186009600
- 2x Microtube Domino | p/n 186009601
- Andrew Alliance Bluetooth Electronic Pipette, 1-ch 300 μL | p/n 186009606
- Andrew Alliance Bluetooth Electronic Pipette, 1-ch 10 μL | p/n 186009769
- Sartorius, Optifit Tips, 0.1-10 μL (x6) | p/n 700013293
- Sartorius, Optifit Tips, 5-350 μL (x4) | p/n 700013297
➤ O-glycopeptide Reduction and Alkylation, 2 Samples, 3 Replicates (Protocol 2/2)
- Tip Insertion System Domino | p/n 186009612
- 2x Microplate Domino | p/n 186009600
- Deepwell Microplate Domino | p/n 186009597
- 96-PCR Plate Peltier+ | p/n 176004584
- Andrew Alliance Bluetooth Microplate Gripper | p/n 186009776
- Andrew Alliance Bluetooth Electronic Pipette, 1-ch 120 μL | p/n 186009765
- Andrew Alliance Bluetooth Electronic Pipette, 8-ch 120 μL | p/n 186009605
- Sartorius, Optifit Tips, 0.5-200 μL (x34) | p/n 700013295
Reagents and Chemicals
- OpeRATOR®, 2000 U by Genovis | p/n G2-OP1-020
- SialEXO®, 2000 U by Genovis (supplied with OpeRATOR)
- Optional - FabRICATOR® (IdeS), 2000 U by Genovis | p/n A0-FR1-020
- 20 mM Tris buffer, pH 7.5
- Guanidinium hydrochloride solution, 8 M, pH 8.5, buffered aqueous solution by Sigma-Aldrich | p/n G7294 or equivalent
- DTT (1,4 Dithiothreitol) by G-Biosciences | p/n 786-077 or equivalent
- Iodoacetamide by G-Biosciences | p/n 786-078 or equivalent
- 6% formic acid
Recommended Consumables
- Fisherbrand™ Premium 1.5 mL microtube | p/n 11926955
- Eppendorf twin.tec® 96-well skirted LoBind® PCR plate | p/n 0030129555
- Axygen® 12-well reservoir, 12-channel trough | p/n RES-MW12-HP
- Waters 350 μL 96-round well collection plate | p/n 186002643
- Optional - Waters polypropylene cap mat for 96-well plate, round wells | p/n 186002483
Learn more about Genovis OpeRATOR® Enzyme
Protocols
Contact info

 This is an open access protocol distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited.
This is an open access protocol distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited. 