or start from open source methods. Learn more about OneLab softwareUse OneLab
NucleoSpin RNA Virus Protocol
This basic method provides the core methodology for translating a workflow into OneLab-executable script(s) as an attempt to fully or semi-automate a specific procedure. It demonstrates the benefits of automation and highlights OneLab capabilities and best practices to promote solution adoption, helping transition from manual to a more automated approach. It can be used alone or serves as a building block for a more complex workflow and is easily adaptable to users' requirements.
Overview
Analysis of Viral Nucleic Acids
Isolation of viral nucleic acids (e.g. viral RNA and DNA) from clinical samples plays a pivotal role in a large variety of molecular tests that have become standard methods in research and diagnostics laboratories, such as real-time (RT)-PCR or next-generation sequencing.
The following features make NucleoSpin RNA Virus ideal for detecting viral nucleic acids :
- Reliable and reproducible isolation of viral RNA and DNA - free of impurities
- Proven for a large variety of viruses including SARS-Cov2, influenza viruses, HCV, HIV, Bird Flu Virus, Blue Tongue Virus
- Carrier RNA included for highest sensitivity in downstream applications
- Fast and convenient protocol - viral RNA within only 30 min
Extraction of Viral RNA for SARS-CoV-2 Detection
The ongoing pandemic of severe acute respiratory syndrome coronavirus 2 (SARS-CoV2) is a major threat to public health. Early detection of infected individuals and understanding the pathway of transmission is one of the main strategies to reduce the further spreading of the disease.
In preparation for COVID-19 molecular testing, laboratories rely on workflows that isolate SARS-CoV-2 RNA from respiratory samples. Multiple labs have already utilized our NucleoSpin RNA Virus kit in their workflows for COVID-19 detection, including the WHO reference laboratory, the National Institute of Health, Thailand (see protocol link below).
High Sensitivity Detection of Viral RNA from Human Saliva
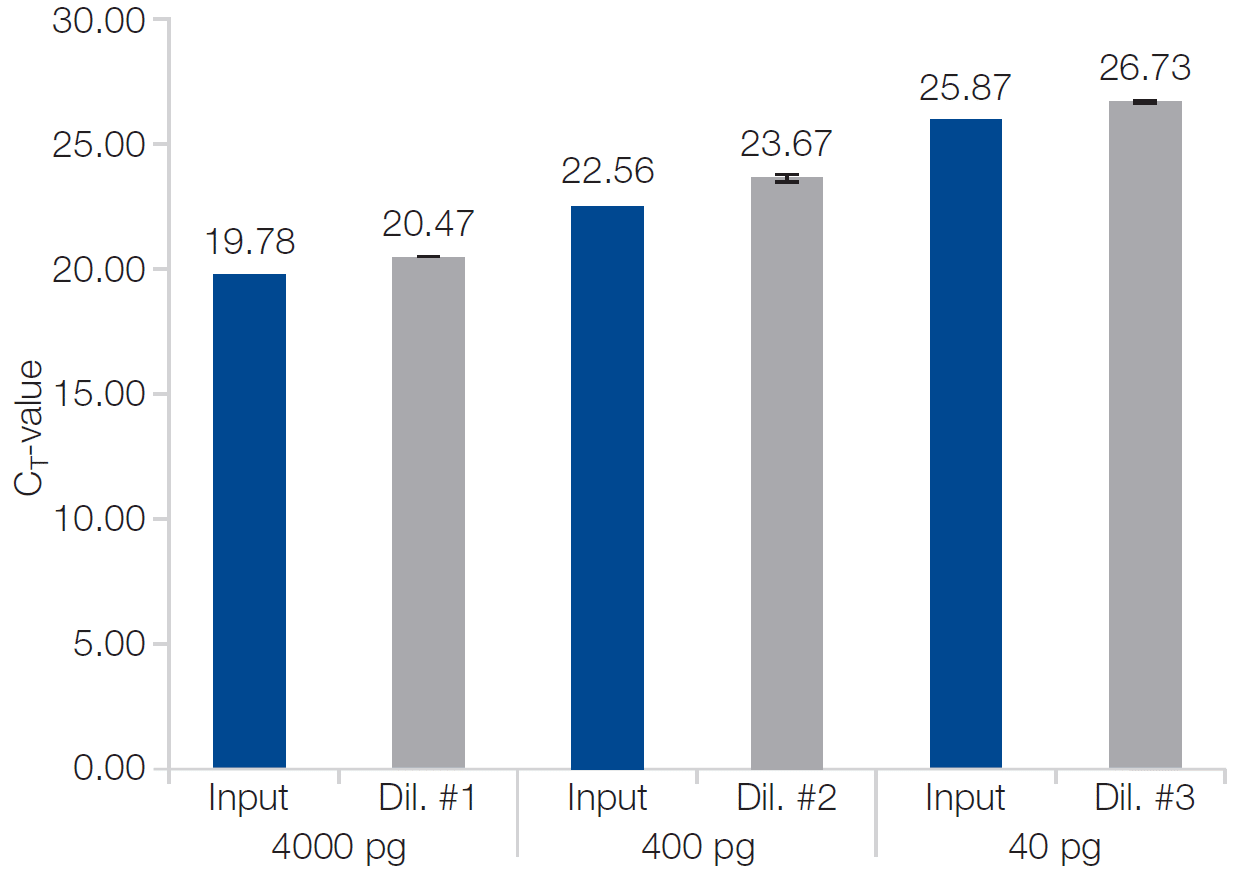
RNA was isolated from saliva (150 µL, duplicates per dilution) using the NucleoSpin RNA Virus kit in combination with the Pipette+ and OneLab system. MS2 bacteriophage RNA was spiked in the respective dilution of the saliva sample. Subsequent qRT-PCR analysis show an excellent recovery rate using the NucleoSpin RNA Virus kit.
Basic Principle of the NucleoSpin RNA Virus Procedure
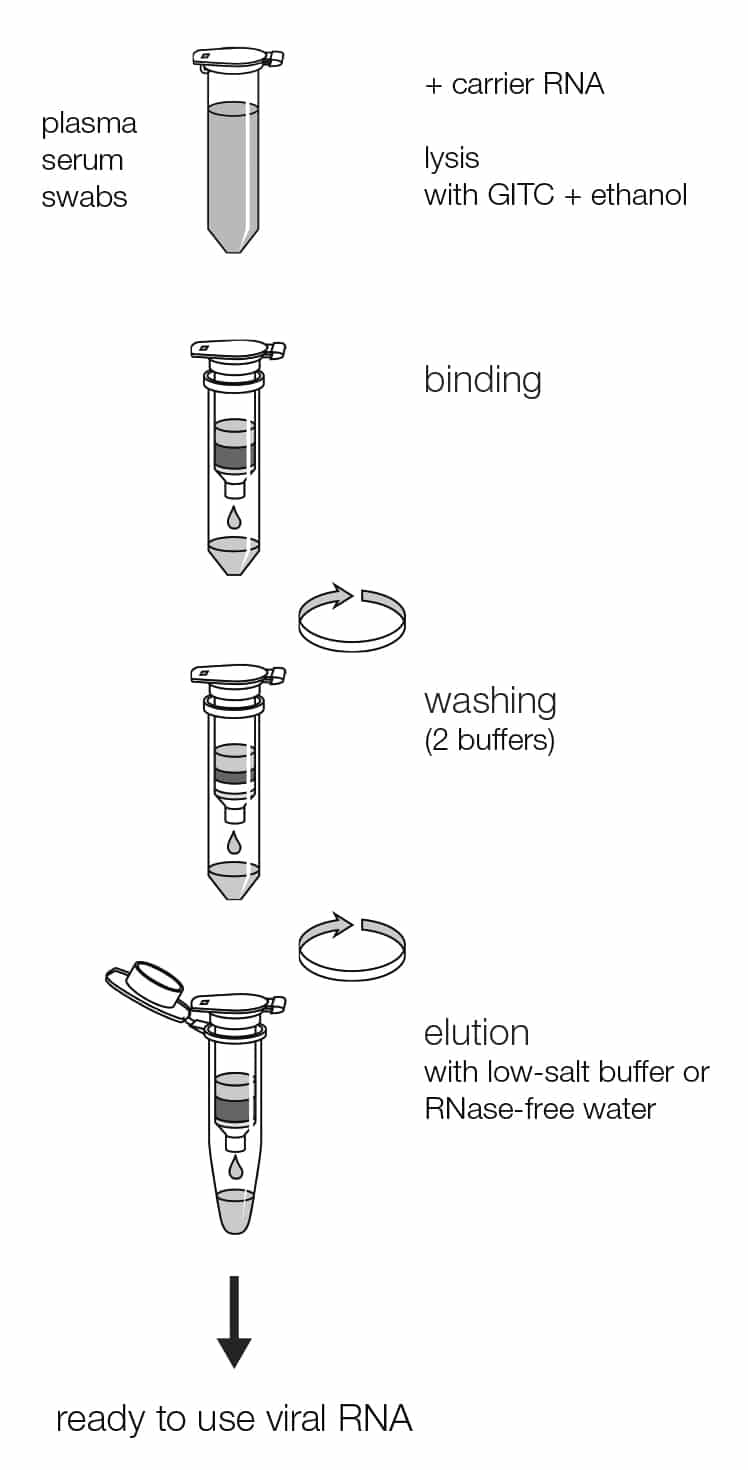
With the NucleoSpin RNA Virus method, RNA viruses are lysed quickly and efficiently by Lysis Buffer RAV1 which is a highly concentrated solution of GITC. DNA viruses (e.g., HBV) are usually more difficult to lyse and require Proteinase K digestion. Lysis buffer and ethanol create appropriate conditions for the binding of nucleic acids to the silica membrane of the NucleoSpin RNA Virus Columns. Carrier RNA improves the binding and recovery of low-concentrated viral RNA. Contamination (potential PCR inhibitors) like salts, metabolites, and soluble macromolecular cellular components are removed in simple washing steps with ethanolic buffers RAW and RAV3. The nucleic acids can be eluted in low salt buffer or water and are ready-for-use in subsequent reactions.
Recommendations for Processing Respiratory Samples
1-Swabs (e.g. naso- or oropharyngeal)
(Dry) Swabs without viral transport media
Rinse swabs with moderate shaking in 400–500 μL of sterile PBS for 30 min to release sample material from the swab (swab heads should be completely submerged in PBS). Transfer 150 μL sample volume of the rinse solution to a suitable reaction container/ tube and proceed with the standard protocol starting with the sample lysis step.
Swabs with viral transport media
Rinse swabs for 30 min with moderate shaking in viral transport media to release sample material from the swab. Transfer 150 μL sample volume to a suitable reaction container/tube and proceed with the standard protocol starting with the sample lysis step.
2-Sputum / Bronchoalveolar Lavage (non-viscous) / Saliva
Non-viscous, clear, and homogenous sputum and bronchoalveolar lavage samples can be used directly for nucleic acid extraction. Transfer 150 μL sample volume to a suitable reaction container/tube and proceed with the standard protocol starting with the sample lysis step.
3-Sputum / Bronchoalveolar Lavage (viscous)
Viscous sputum and bronchoalveolar lavage samples should be liquefied before subjecting them to the nucleic extraction procedure. Transfer 150 μL sample volume to a suitable reaction container/tube. Add 600 μL of lysis buffer RAV1 to the sample and incubate at 70 °C for 10 min with moderate shaking. Check if the sample is liquefied, allow the sample to cool down, and proceed with the binding step.
If the sample is not liquefied after the heat incubation, follow CDC's recommended guidelines for processing sputum samples. Transfer 150 μL of the liquefied sample to a suitable reaction container/tube and proceed with the standard protocol starting with the sample lysis step.
Order NucleoSpin RNA Virus for purification of viral RNA from cell-free fluids
Find more information about the NucleoSpin RNA Virus procedure in our manual
About MACHEREY-NAGEL ́s latest references for Covid-19 and viral nucleic acid isolation
Protocols

Contact info

 This is an open access protocol distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited.
This is an open access protocol distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited. 