or start from open source methods. Learn more about OneLab softwareUse OneLab
MTBE Lipid Extraction
This basic method provides the core methodology for translating a workflow into OneLab-executable script(s) as an attempt to fully or semi-automate a specific procedure. It demonstrates the benefits of automation and highlights OneLab capabilities and best practices to promote solution adoption, helping transition from manual to a more automated approach. It can be used alone or serves as a building block for a more complex workflow and is easily adaptable to users' requirements.
Overview
Lipid Extraction from Biological Matrices
Lipids are a diverse class of biomolecules involved in many metabolic processes and have therefore also been identified as being markers of different metabolic diseases. Due to the variety of physiochemical properties that the lipids possess, being able to extract the broad collection of subclasses from biological matrices is essential.
Lipid extraction using methyl tert-butyl ether (MTBE) has previously been shown to provide the broadest coverage of lipid class extraction from plasma, but being a liquid-liquid extraction method, numerous pipetting steps and phase separation are required (Figure 1). When comparing this to the more widely used isopropanol extraction method which has fewer steps, this can put a strain on resources and can lead to the introduction of errors impacting the quality and ultimately the confidence of the results.
Furthermore, the analysis of lipids from samples obtained from large-scale population studies can involve sample preparation of hundreds to thousands of samples in order to provide adequate statistical power. This in turn is time-consuming and requires close management of the logistics of study sample preparation to maintain sample integrity.
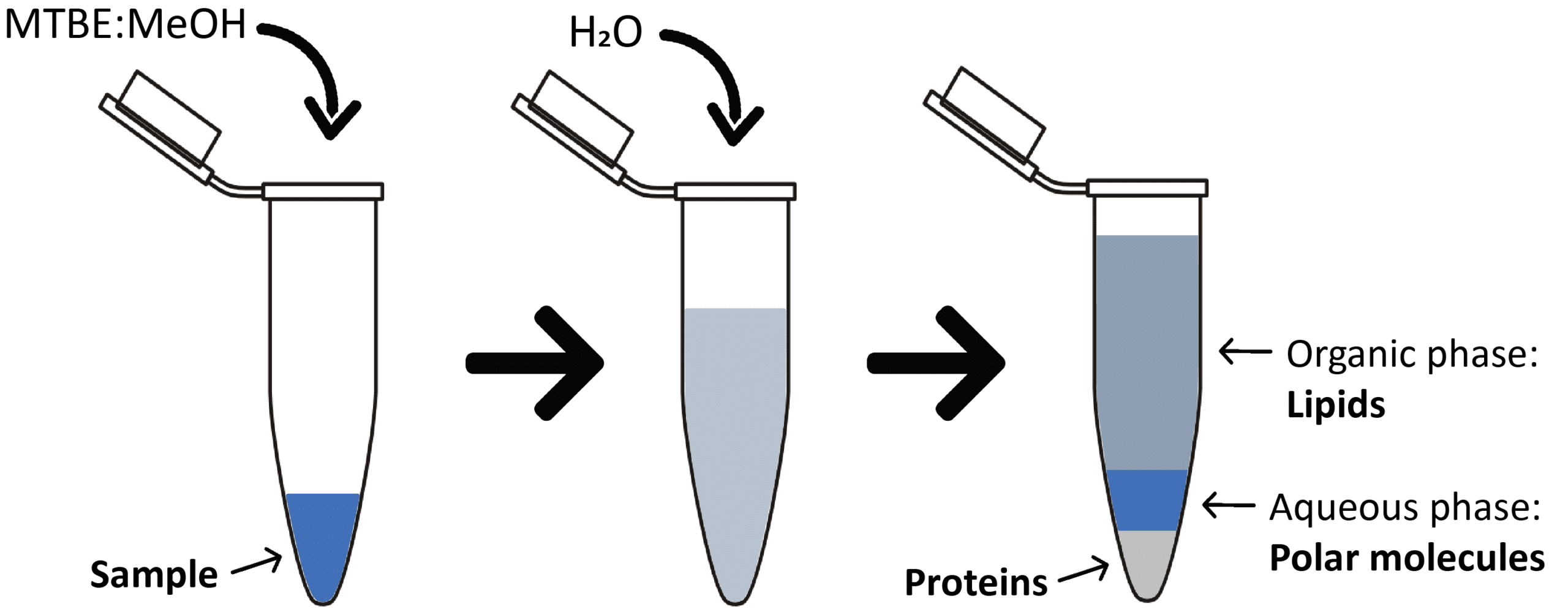
Figure 1: MTBE extraction method schematic.
Andrew+ Automated MTBE Extraction Protocol
To assist in managing the extraction of lipids using the MTBE methodology, we outline here an automated pipetting protocol using the Andrew+ Pipetting Robot. The main choke point of this extraction procedure is the numerous repetitive pipetting steps. This OneLab protocol's main goal is in addressing this issue. All pipetting steps are performed by the Andrew+ robot, which includes the addition of methanol, MTBE, and water, as well as the final aliquoting of the two phases post-extraction. These pipetting steps are interspersed with several manual operator actions which include vortex mixing/shaking, centrifugation, and the incubation step to ensure protein precipitation and lipid extraction.
To perform manually, the pipetting steps for the whole extraction procedure would take an estimated 1 hour and 10 minutes. Conversely, when using the Andrew+ robot, pipetting was reduced to 40 minutes increasing the throughput of the extraction procedure in addition to enabling the operator to focus on other tasks. Furthermore, the protocol aliquots the lower aqueous phase in addition to the upper organic lipid phase which can be retained and analyzed additionally for small polar molecule analysis. This provides a more comprehensive extraction method for lipidomic and metabolomic profiling negating the need for additional samples for further extractions.
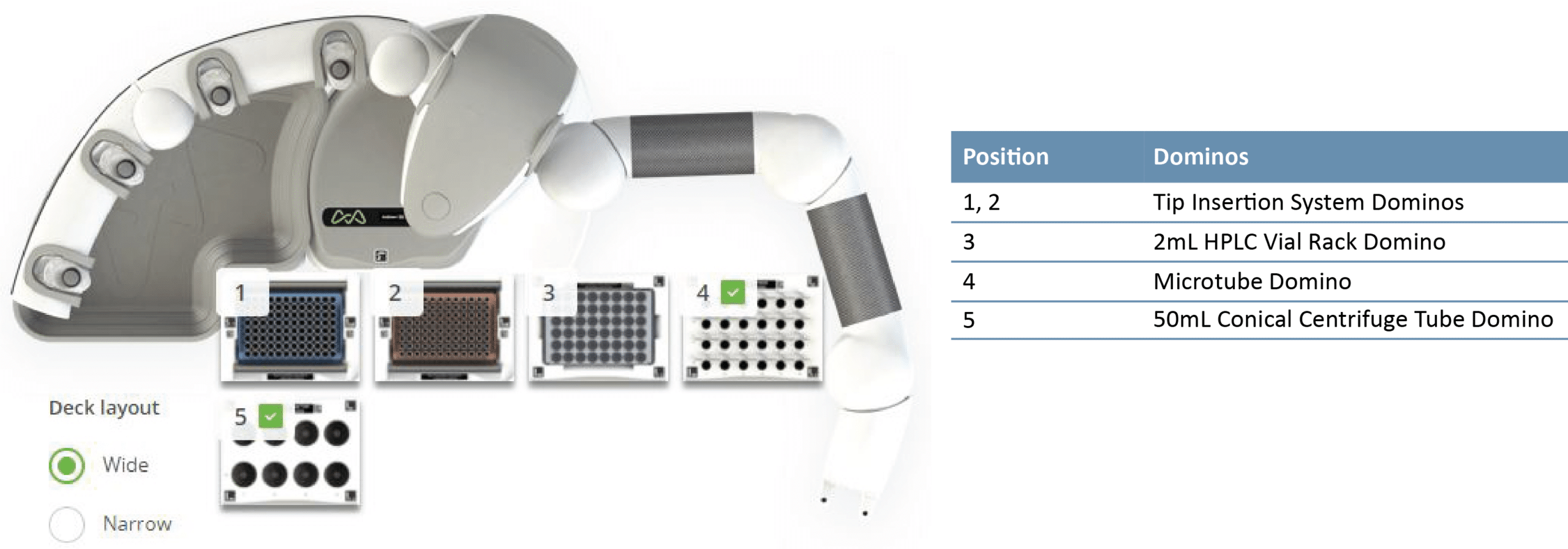
Figure 2: Andrew+ Domino configuration for MTBE extraction protocol (n = 24 study samples).
MTBE LLE Protocol Steps
The workflow in Figure 3 describes the protein precipitation, liquid-liquid extraction, and aliquoting of both organic and aqueous phases of biological matrices for the subsequent analysis by LC-MS/MS.
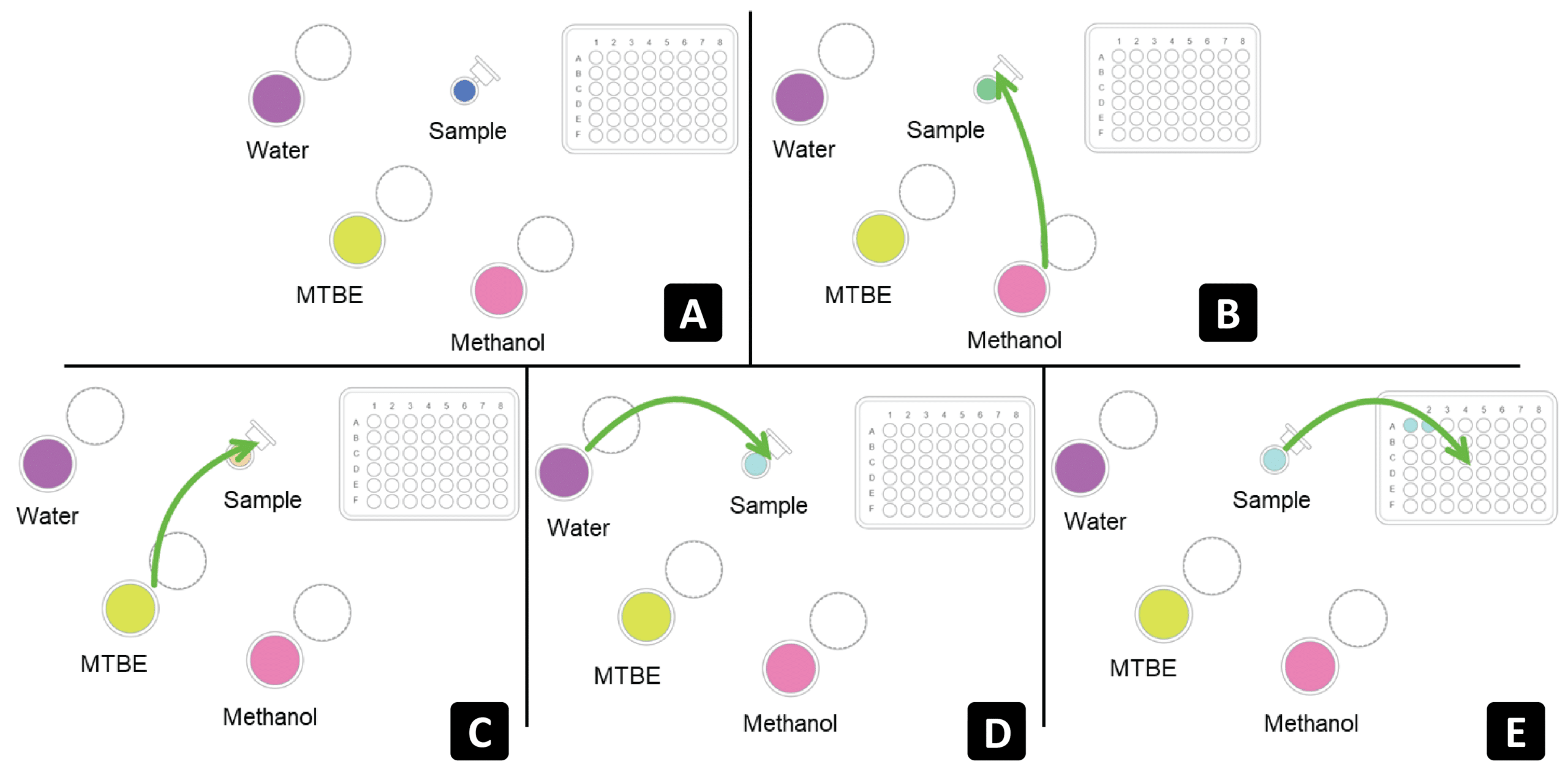
Figure 3: Schematic outline of Andrew+ pipetting steps for the MTBE lipid LLE extraction method.
➤ Step A
Manually, 100 μL of plasma is individually aliquoted into 1.5 mL microtube tubes (n = 24) and are then transferred to the Microtube Domino and left uncapped.
➤ Step B
To each sample, 200 μL of methanol is added using the Andrew+ Pipetting Robot.
➤ Step C
Next, MTBE is added (800 μL) to each of the samples before being capped manually. Once sealed, each sample is vortex mixed briefly (10 sec per sample) before all samples are incubated at 2-8°C for 1 hour to ensure full protein precipitation and lipid extraction.
➤ Step D
Following incubation, samples are returned to the Andrew+ robot un-capped and 300 μL of deionized water is then added to each sample. Following this, samples are vortex mixed again (10 sec per sample) and then centrifuged at 13,000 rpm for 10 minutes to cause phase separation and pellet precipitated proteins.
➤ Step E
After centrifugation, each study sample is returned to the Microtube Domino where the top phase of the supernatant is removed and transferred to a Waters total recovery vial for lipid analysis. The lower aqueous phase is then transferred to a separate total recovery vial for analysis of small molecules and additional polar lipids.
ORDERING INFORMATION
Andrew+ System Components: Dominos, Electronic Pipettes & Tips
- 50mL Conical Centrifuge Tube Domino | p/n 186009614
- 2mL HPLC Vial Rack Domino | p/n 186010091
- Microtube Domino | p/n 186009601
- 2x Tip Insertion System Domino | p/n 186009612
- Andrew Alliance Bluetooth Electronic Pipette, 1-ch 300 μL | p/n 186009606
- Andrew Alliance Bluetooth Electronic Pipette, 1-ch 1000 μL | p/n 186009766
- Sartorius, Optifit Tips, 10-1000 μL
- Sartorius, Optifit Tips, 5-350 μL
Consumables
- Corning® 50 mL conical centrifuge tube | p/n 430290
- Eppendorf 1.5 mL Safe-lock tube | p/n 0030120086
- Waters 1 mL LC/GC screw-top vial in 48x holder | p/n 186000385C
Protocols

Contact info

 This is an open access protocol distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited.
This is an open access protocol distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited. 