or start from open source methods. Learn more about OneLab softwareUse OneLab
Matrix-Matched Pesticide Standard Curve Preparation

This basic method provides the core methodology for translating a workflow into OneLab-executable script(s) as an attempt to fully or semi-automate a specific procedure. It demonstrates the benefits of automation and highlights OneLab capabilities and best practices to promote solution adoption, helping transition from manual to a more automated approach. It can be used alone or serves as a building block for a more complex workflow and is easily adaptable to users' requirements.
Overview
Pesticide Analysis in Agricultural Commodities
Matrix effects are a common concern in pesticide analysis, which can compromise detection and quantification quality. To improve the efficiency of traditional methods, Anastassiades et al. introduced a sample preparation approach known as QuEChERS (Quick, Easy, Cheap, Effective, Rugged, and Safe). This procedure uses a single extraction in acetonitrile and requires a small amount of sample (10-15 g). The resulting acetonitrile extract is typically analyzed directly by GC-MS/MS or LC-MS/MS after proper dilution.
Challenges in Sample Preparation for Matrix-Matched Standard Curve Generation
The initial mobile phase conditions for a typical LC-MS/MS analysis is water/acetonitrile (9:1). Sample solvent with stronger elution strength than initial conditions of the mobile phase causes peak distortion, peak broadening, and earlier elution due to less effective retention. A proper dilution after QuEChERS extraction is essential to ensure successful and reproducible LC-MS/MS analysis. Moreover, it is also recommended to pre-wet the tips when handling volatile solvents like acetonitrile, methanol, isopropanol, etc., to reduce dripping. All these considerations further increase the number of pipetting tasks required in a typical workflow.
While often not difficult, preparing these samples is repetitive, time-consuming, and requiring consistency to ensure the reliable performance of the analytical method. This makes it ideal for incorporating lab automation, thus allows the analyst to do other tasks, streamlines the sample preparation process, reduces the potential for human error, and ensures consistent analytical method performance.
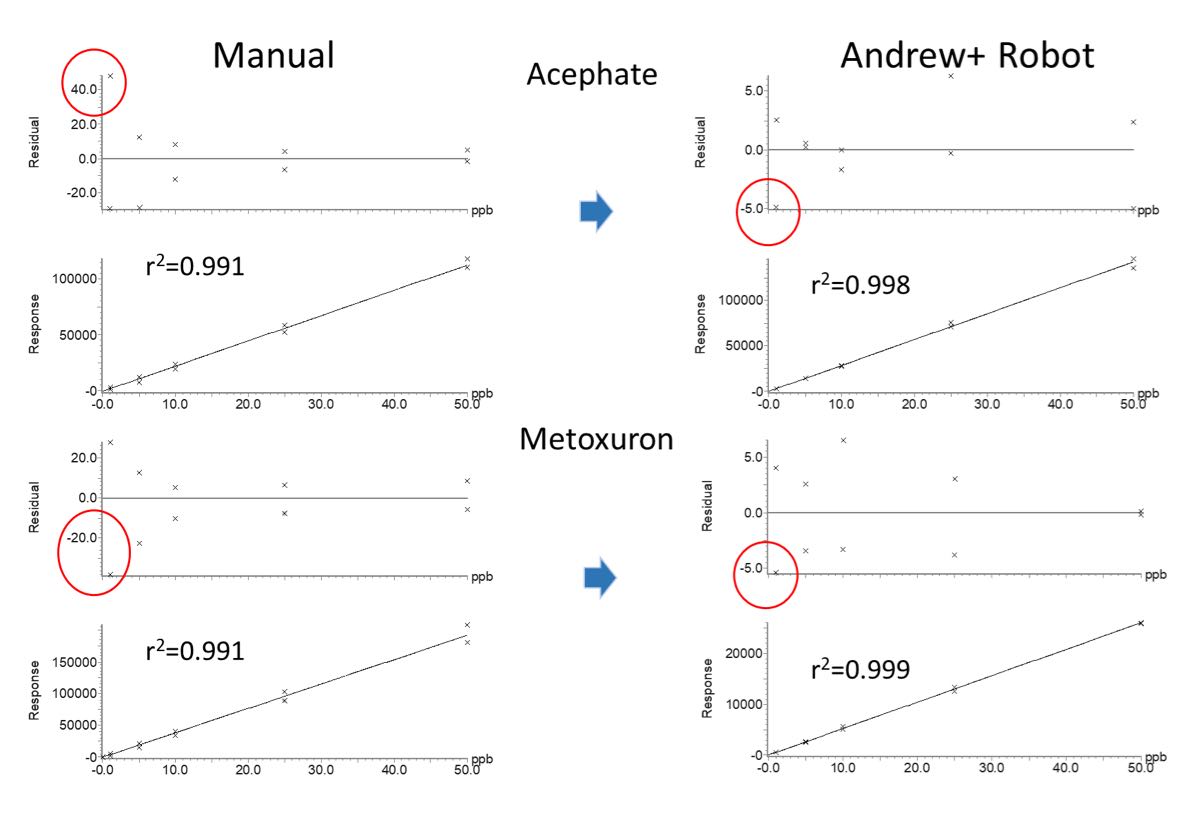
Figure 1: Typical calibration graphs generated from the analysis of matrix-matched standards prepared in duplicate
Assay notes
This protocol was developed to generate seven calibration levels plus a blank. Column 1 consists of the standard stock solutions at ten times the concentration. Columns 2 and 3 are duplicates of matrix-matched calibration standards. Columns 4 and 5 are duplicates of calibration standards prepared in solvent. By preparing two formats of calibration curve (matrix vs. solvent), it allows the user to investigate and understand the matrix effect.
The standard stock solutions are first prepared in Column 1 to yield the concentration at 10, 50, 100, 250, 500, 750 and 1000 ppb. Matrix-matched calibration curve is prepared by adding 10 μL standards + 10 μL matrix + 80 μL water to give a total of 100 μL volume. 10 μL of matrix is replaced with acetonitrile when preparing calibration standards in solvent.
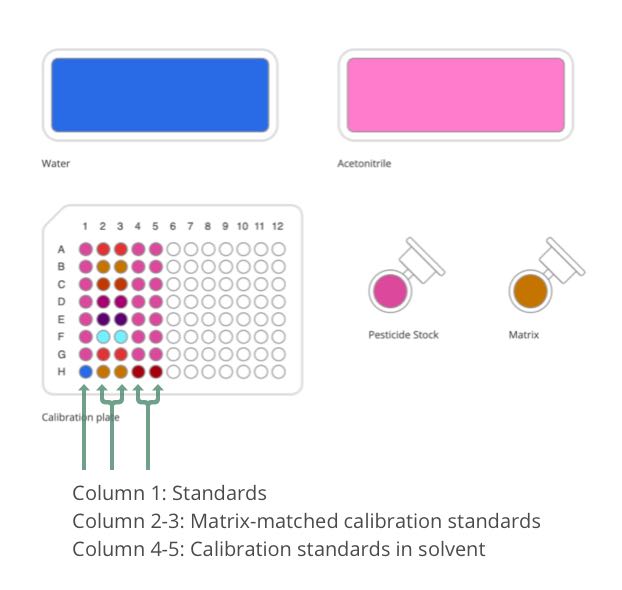
Figure 2: Labware and solutions required for matrix-matched calibration curve preparation
Protocol options
Protocol 1 - Matrix-matched calibration curve only
This option allows the user to build a matrix-matched calibration curve with the proper dilution suitable for typical LC-MS/MS analysis.
Protocol 2 - Matrix-matched calibration curve + calibration standards prepared in solvent
This option allows the user to investigate and understand the matrix effect.

Figure 3: Andrew+ OneLab deck set up for the calibration standards preparation. Samples and reagents locations on Andrew+ deck: [1] Tip Insertion System Domino, [2] Deepwell Microplate Domino, [3] Microtube Domino, [4] 8-Channel Reservoir Domino
Protocol specifications
Apple Matrix Pesticide Calibration - Protocol 1:
- Estimated time of execution: 17m 4s
- Hands-on time: 0s
- Tip consumption:
- 41× 10-300 μL tips
Apple Matrix Pesticide Calibration - Protocol 2:
- Estimated time of execution: 18m 37s
- Hands-on time: 0s
- Tip consumption:
- 66× 10-300 μL tips
Ordering information
Andrew+ System Components: Dominos, Devices, Electronic Pipettes & Tips
- Andrew+ Pipetting Robot
- OneLab software
- Tip Insertion Domino | p/n 186009612
- Deepwell Microplate Domino | p/n 186009597
- Microtube Domino | p/n 186009601
- 8-Channel Pipette Reservoir Domino | p/n 186009613
- Andrew Alliance Bluetooth Electronic Pipette, 1-ch 300 μL | p/n 186009606
- Andrew Alliance Bluetooth Electronic Pipette, 8-ch 300 μL | p/n 186009607
Recommended consumables
- Waters 20 Pesticide Mix Standard | p/n 186006348
References
- Waters Application Note 720007433EN - Automating Preparation of Matrix-Matched Standards for Pesticide Residue Analysis Using the Andrew+ Pipetting Robot (https://www.waters.com/content/dam/waters/en/app-notes/2021/720007433/720007433-en.pdf)
Protocols

Contact info

 This is an open access protocol distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited.
This is an open access protocol distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited. 