or start from open source methods. Learn more about OneLab softwareUse OneLab
Magnetic His-Protein Purification

This example method provides a freely adjustable framework for measuring the adaptability of the OneLab environment to workflows from different application fields. It helps to understand various nuances of the code-free, universal protocol designer and provides general indications as to the feasibility of a project. Scripts generally require fine adjustment to correct for variables and support specific labware implementation.
Overview
Recombinant Proteins and Metal Affinity-Based Separation
Recombinant proteins are produced by heterologous expression of recombinant DNA (coding gene) in host organisms (e.g. mammalian cells, insect cells, yeast, bacteria). Recombinant proteins are involved in wide applications including the development of biopharmaceuticals for therapeutic use, enzymes for industrial applications and bioassay development for diagnostics. They also serve as research tools and reagents in proteomics research for studying gene function, characterization of protein structure, protein interactions, drug screening, and antibody development.
The economical production of recombinant proteins requires not only an efficient expression system but also the implementation of an optimized and reliable purification method that subsequently enables their study and/or application. Strategies for protein purification are mostly focused on chromatography purification which separates target proteins according to the difference between their properties and those of other substances present in the sample mixture. The most widely used method of purification is affinity chromatography (AC) which separates target proteins based on their reversible interaction with a specific ligand attached to an insoluble matrix. The immobilized ligand can be either a biological entity, for example, Protein A for purification of monoclonal antibodies, or a chemical compound, for example, proteins containing natural surface-exposed histidine residues interacting with nickel ions (Ni2+). This intrinsic metal-binding ability of some amino acids (e.g. histidine, cysteine or tryptophan) is the basis of the Immobilized Metal Affinity Chromatography or IMAC where proteins expressing these residues on their surface interact with specific metal ions bound by chelation to the solid support. This strategy was also exploited to purify recombinant proteins with engineered histidine-tag sequence in a single-step procedure. Thanks to the DNA recombinant technology, the fusion strategy has facilitated the purification and identification of recombinant proteins by introducing a specific poly-histidine affinity tag (6 to 10 consecutive histidine or His residues) to the protein sequence, leading to the expression of a histidine-tagged fusion protein. His-tagged fusion proteins are then selectively purified by interaction between the histidine and several divalent cations such as Ni2+, Cu2+, Zn2+, and Co2+. Other affinity tags exist (e.g. glutathione S-transferase, GST tag) but poly-histidine is the most commonly used tag because it has a short sequence, is non-immunogenic and does not affect the folding or biological activity of the tagged protein. Moreover, the histidine tag confers high affinity to recombinant proteins which bind tightly to the stationary phase, while other cellular proteins in a crude sample extract (e.g. cellular lysate) will not bind or will bind weakly. This feature enables purification of tagged recombinant proteins at a high purity level (up to 95%) and recovery rate in a one-step purification under both native and denaturing conditions.
Chromatography methods are often time-consuming and costly as they require optimization to meet the physicochemical properties of the sample and the required purification level. An adequate purification procedure should ensure a certain balance between key parameters, including speed of purification, recovery rate and capacity, on which will depend the outcome. In optimal conditions, a purification method should be able to eliminate host contaminants and efficiently separate intact target proteins (solubility, structure integrity and biological activity should be preserved), resulting in high purity products and enough yield. To assess analytical conditions and accelerate biological discovery, it is critical to apply simple and rapid methods for purification of new recombinant proteins that require minimal optimization and can be scalable and easily integrated with benchtop lab automation.
Purification of His-Tagged Proteins using PierceTM Ni-NTA Magnetic Agarose Beads
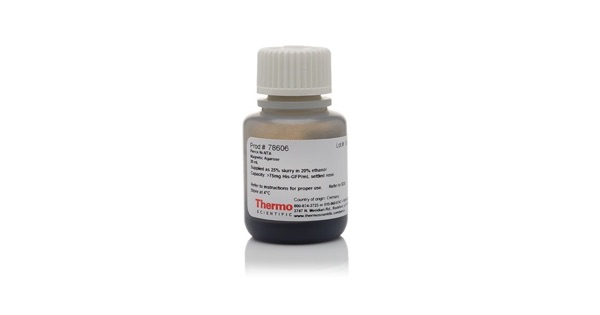
This protocol provides a fast and convenient method to efficiently purify recombinant His-tagged proteins in a single-step procedure using magnetic separation. The strategy combines the His-Ni2+ binding ability and magnetic feature of proprietary beads to selectively separate recombinant His-tagged proteins from a sample mixture (e.g. cell lysate) in 1.5 mL low protein binding microcentrifuge tubes.
Ni-NTA magnetic agarose beads (10-40 μm) are made of highly cross-linked and magnetite-embedded agarose coupled to tetradentate nitrilotriacetic acid (NTA) chelating ligand, which in turn is charged with divalent nickel ions (Ni-NTA). The Ni-NTA resin coordinates with the histidine side chains enabling selective purification of His-tagged proteins (Figure 1). The binding capacity of the Ni-NTA magnetic agarose beads can go up to 75 mg of His-tagged protein per 1 mL of settled beads (4 mL of 25% bead slurry). The volume of beads can then vary depending on the predicted amount of protein in the sample and the type of magnetic apparatus, for instance, for small-scale purification, 25 μl of settled beads (100 μl of 25% bead slurry) is used to purify between 1 mg to 1.9 mg of His-tagged protein.
This protocol is very useful for small-scale affinity purification of recombinant His-tagged proteins. It is also useful for screening for optimal expression and purification conditions to be used in larger-scale purification using chromatography techniques. The non-aggregating, superparamagnetic Ni-NTA agarose beads provide high performance and exceptional uniformity for both manual and automated benchtop purification applications using an appropriate magnetic stand. They are optimized to ensure low non-specific binding and high purity of the isolated His-tagged proteins in both native or denaturing conditions.
The protein yield and purity delivered by the protocol are dependent upon the expression level, conformation and solubility characteristics of the recombinant fusion protein. Therefore, it is crucial to optimize these parameters, for example, by performing a small-scale run test to estimate the adequate expression level and determine the solubility of the recombinant His-tagged protein.
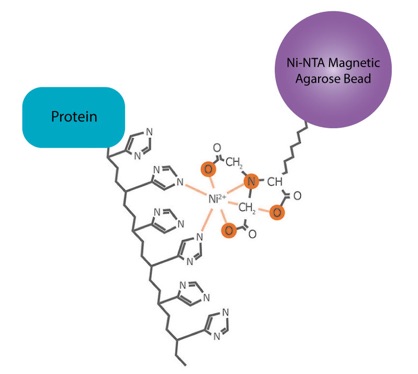
Figure 1: A schematic illustration showing the mechanism of interaction between the Ni-NTA resin and the poly-histidine tag. The NTA chelator contains four metal-binding sites, three oxygen atoms and one nitrogen atom, that allow for strong binding and consequently low metal ion leaching. The nickel ion (Ni2+) coordinates with the side chains of adjacent histidine residues enabling rapid and selective one-step affinity purification of His-tagged proteins.
Protocols
Contact info

 This is an open access protocol distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited.
This is an open access protocol distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited. 