or start from open source methods. Learn more about OneLab softwareUse OneLab
KAPA DNA Library Quantification
This example method provides a freely adjustable framework for measuring the adaptability of the OneLab environment to workflows from different application fields. It helps to understand various nuances of the code-free, universal protocol designer and provides general indications as to the feasibility of a project. Scripts generally require fine adjustment to correct for variables and support specific labware implementation.
Overview
Accurate quantification of DNA libraries is a critical step in the Illumina NGS workflow allowing for maximizing yield and achieving high-quality sequencing data. Each library must be properly quantified before pooling and/or loading onto a flow cell to ensure equimolar amounts of indexed libraries in multiplexed applications and an optimal cluster density. Underestimation of library concentration may result in higher than desired cluster density (over-clustering), which hinders cluster resolution, while overestimation may generate lower cluster density (under-clustering).
Absolute Quantification of DNA Libraries using Quantitative Real-Time PCR
Individual and pooled libraries need to be prepared at specific molarity to ensure compatibility with the sequencing platform and maximize sequencing capacity. Usual methods for nucleic acid quantification including capillary gel electrophoresis (Bioanalyzer), UV spectrophotometry (NanoDropTM instrument), and fluorometry (Qubit® fluorometer) measure total DNA concentrations including non-amplifiable DNA. Furthermore, the spectrophotometer is very sensitive to contaminants such as organic solvents (e.g. ethanol), used during library purification and clean-up steps, that absorb in the UV spectrum, while quantification using an electrophoretic assay may have high inherent variation. The most accurate and sensitive method to quantify libraries is quantitative real-time PCR, which measures only the number of amplifiable DNA molecules with complete sequencing adaptors at both ends, even in very dilute libraries.
KAPA Library Quantification Kit
The KAPA Library Quantification Kit provides a reliable qPCR-based solution for the absolute quantification of individual libraries or indexed library pools prepared for sequencing on Illumina platforms. It measures specifically the concentration of all sequencing-competent DNA molecules flanked by adapters containing the Illumina P5 and P7 flow cell oligo sequences. The kit comprises all the reagents required to achieve an accurate concentration measurement:
- The KAPA SYBR® FAST DNA qPCR Master Mix (2X): It includes a novel KAPA SYBR® FAST DNA Polymerase optimized to enable high-performance real-time qPCR using the SYBR® Green I dye chemistry. The ability of this enzyme to amplify diverse DNA fragments in a complex population with similar efficiency makes it ideally suitable for use with universal DNA standard for quantification of all types of Illumina libraries containing fragments up to 1 kb in length with a broad range of concentrations (>0.0002 pM) and GC content. The master mix is formulated with various passive dyes for compatibility with different qPCR instruments. Passive dyes are used in qPCR reactions as an internal reference to normalize for non-PCR-related fluorescence signal variation.
- Library Quantification DNA Standards numbered 1 to 6: The pre-diluted set of DNA standards consists of a 10-fold dilution series of a linear, 452 bp DNA template flanked by qPCR primer binding sites.
- Library Quantification Primer Premix (10X): It contains a set of two primers complementary to the Illumina P5 and P7 flow cell oligo sequences.
Principle of the KAPA Library Quantification Assay
Pre-diluted DNA standards and freshly diluted library samples are prepared in triplicate and analyzed by qPCR. Libraries are diluted to fall within the dynamic range of the KAPA Library Quantification assay (20 – 0.0002 pM). Initial library dilutions are based on estimations from experiments conducted with similar types of libraries or prepared using similar workflows and/or on concentration information obtained with other quantification methods during library construction and quality control (e.g. Qubit® or Bioanalyzer instruments). Two controls are run in parallel: The no-template control (NTC) to detect potential contamination introduced during reaction setup, and an internal dilution control named “Standard 0” to detect gross errors that occurred during sample dilution. Standard 0 is a quality-controlled 200 pM solution of the same linear, 452 bp dsDNA fragment used for the DNA standards. It undergoes the same preparation process as the library samples - the same dilutions and number of replicate reactions are set up.
Determination of Library Concentrations
The average Cq values of DNA standards 1 to 6 are plotted against log10(concentration in pM) to generate a standard curve (Figure 1). The standard curve is used to convert the average Cq value for each library dilution to concentration (in pM). Then, a size-adjustment calculation is performed to compensate for any difference between the length of the DNA Standard (452 bp) and the average fragment length of library samples. The size-adjustment calculation consists of multiplying the concentration of the library dilution derived from the standard curve with the ratio between the size of the DNA Standard (452 bp) and the average fragment size for that library. Finally, size-adjusted concentrations for each library are corrected for dilution and an average concentration of the undiluted library is calculated. This working concentration is used to prepare an appropriate dilution of the library and proceed with the loading of the flow cell and cluster amplification.
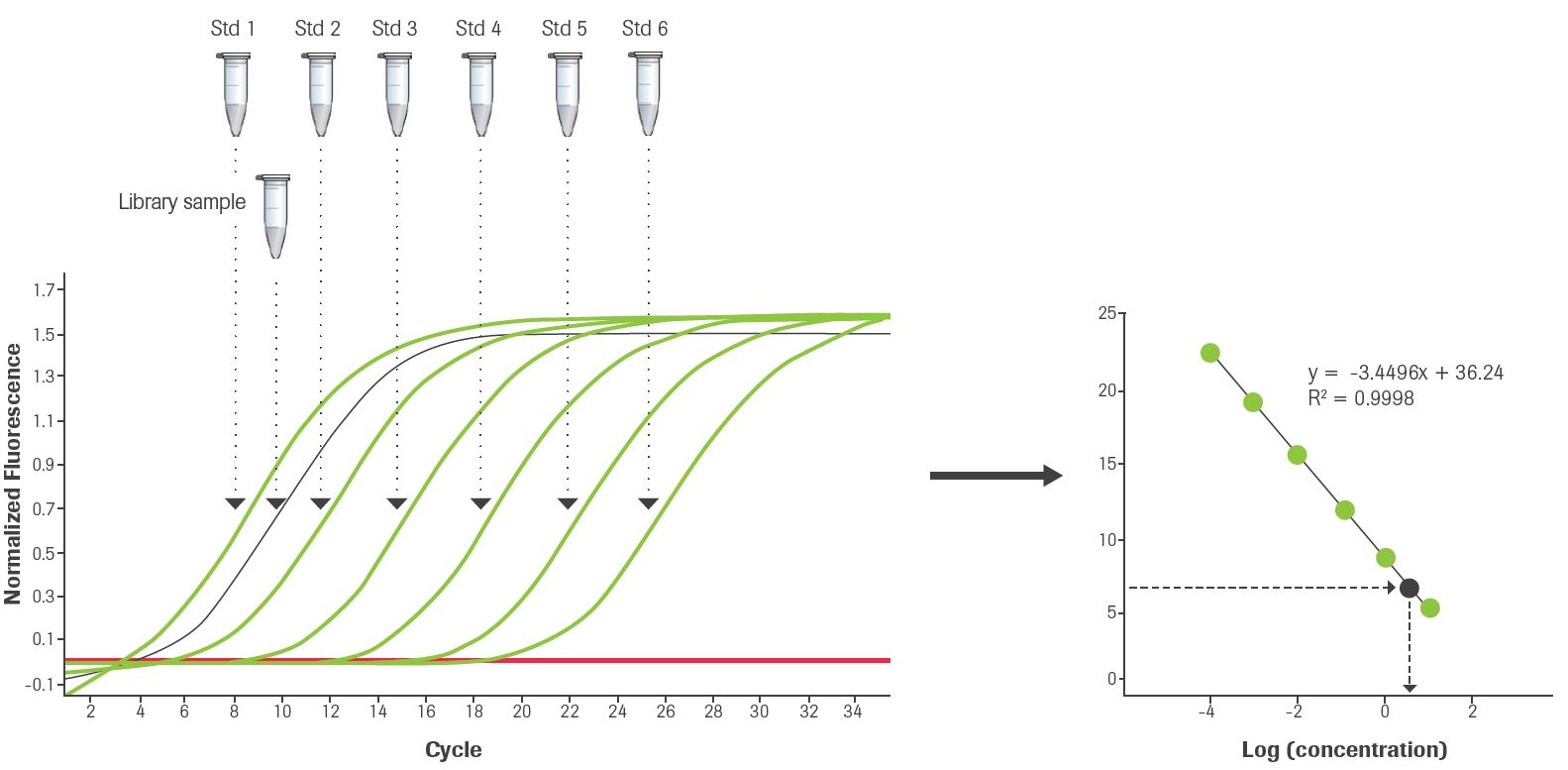
Figure 1: Schematic representation showing the qPCR amplification curves of DNA standards 1-6 and a diluted library sample, and how library concentration is determined by inference from the standard curve generated using the six DNA Standards.
The sensitivity of qPCR makes it very vulnerable to variations arising from various sources notably inaccuracies introduced by inconsistent pipetting technique and cross-contamination. Therefore, successful and reliable qPCR-based library quantification is highly dependent on accurate liquid handling during the preparation of standards, dilution of DNA libraries, and qPCR setup to ensure efficient amplification of all adapter-ligated molecules.
Protocols

Contact info

 This is an open access protocol distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited.
This is an open access protocol distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited. 