or start from open source methods. Learn more about OneLab softwareUse OneLab
DNA Sequencing Library Preparation
This example method provides a freely adjustable framework for measuring the adaptability of the OneLab environment to workflows from different application fields. It helps to understand various nuances of the code-free, universal protocol designer and provides general indications as to the feasibility of a project. Scripts generally require fine adjustment to correct for variables and support specific labware implementation.
Overview
DNA library preparation typically involves two phases: the fragmentation of genomic DNA and the ligation of unique adapter sequences required for sequencing cluster generation. The protocol described here uses a process named “on-bead tagmentation” that combines DNA fragmentation and ligation reactions into a single step using beads carrying modified transposomes. These modified bead-linked transposomes, known as BLT, catalyze simultaneously DNA cleavage and the addition of Read1 and Read2 sequencing primers (Figure 1). The improved bead-linked reaction generates highly uniform insert sizes (300-350 bp) regardless of the DNA input amount (1-500 ng) and genome size. Moreover, it includes an integrated DNA normalization process mediated by BLT saturation that delivers a consistent normalized yield of tagmented DNA within the DNA input range of 100-500 ng, thus eliminating the need for downstream quantification and normalization of individual libraries prior to pooling.
Following post tagmentation cleanup, a reduced-cycle PCR amplification incorporates specific combinations of indexes flanked by P5 and P7 adapters for library hybridization on the sequencing flow cell (Figure 1). Amplified libraries are finally purified using a double-sided bead purification procedure to select sequencing-ready fragments with the target insert sizes (500-600 bp). Libraries can then be pooled and subsequently diluted to the starting concentration required for the sequencing system being used.
The overall advantage of this bead-based technology is the delivery of uniform genome coverage together with minimal bias, thus generating high-quality sequencing-ready DNA libraries. This protocol offers high flexibility by supporting a broad range of DNA input (1-500 ng) and is suitable for a variety of NGS applications, from human whole-genome sequencing and large, complex genomes to PCR amplicons, plasmids, and small microbial species.
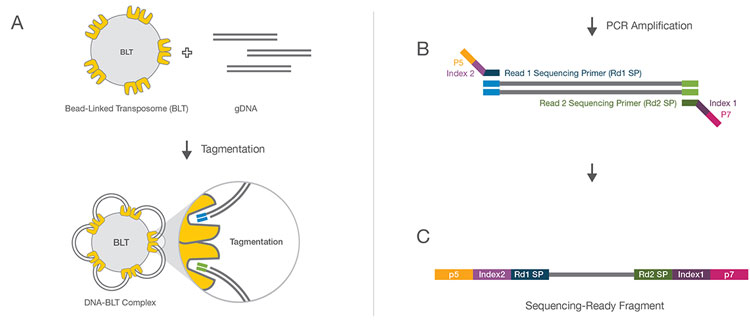
Figure 1: Nextera Bead-Linked Transposome Technology. A) Bead-Linked Transposomes (BLT) drive the simultaneous fragmentation of genomic DNA and the addition of sequencing primers.
B) Reduced-cycle PCR amplification allows for the incorporation of specific index-adapter sequences, thus generating sequencing-ready fragments.
C) Sequencing-ready fragments are finally purified and pooled prior to initiation of sequencing.
Original image from product page “Nextera DNA Flex Library Prep Kit”, in Supporting Data and Figures, Illumina.
Watch the video tutorial providing a comprehensive description of a complete workflow from sample preparation using the "Nextera DNA Flex library prep" approach and cluster generation to sequencing and data analysis
Explore Illumina Nextera DNA Flex Library Prep Kit
Protocols
Contact info

 This is an open access protocol distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited.
This is an open access protocol distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited. 