or start from open source methods. Learn more about OneLab softwareUse OneLab
Detection of 2019-nCoV by RT-qPCR
This example method provides a freely adjustable framework for measuring the adaptability of the OneLab environment to workflows from different application fields. It helps to understand various nuances of the code-free, universal protocol designer and provides general indications as to the feasibility of a project. Scripts generally require fine adjustment to correct for variables and support specific labware implementation.
Overview
The ongoing outbreak of the emerged novel coronavirus termed severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) has raised several challenges for public health and clinical testing laboratories especially in relation with the performance and reliability of results delivered by real-time RT-PCR, which is widely used as a testing method for the identification of virus-infected individuals. In a such international health emergency, the priority is to ensure that the diagnostic method is robust enough to provide specialists with accurate results in a timely manner thereby facilitating the interventions of public health authorities to control and reduce the spread of the disease through application of appropriate treatment or other patient management decisions.
In addition to quality, lab capacity also needs to increase to cope with the growing number of cases requiring testing and monitoring. Therefore, the implementation of liquid handling automation becomes crucial to scale up the number of tests that can be completed each day while ensuring data accuracy. Furthermore, liquid handling automation is an important asset in proficient diagnostic laboratories as requisite optimization and validation of PCR-based assays need to be completed over very short timescales to accelerate the delivery of new diagnostic tests.
Diagnostic Test for Detection of SARS-CoV-2 Virus developed by Centers for Disease Control and Prevention (CDC)
The CDC 2019 Novel Coronavirus (2019-nCoV) Real-Time Reverse Transcriptase (RT)–PCR Diagnostic Panel a molecular in vitro diagnostic test for the qualitative detection of nucleic acid from the 2019-nCoV in upper and lower respiratory specimens collected from individuals who meet 2019-nCoV clinical and/or epidemiological criteria. The product uses polymerase-based nucleic acid amplification technology and comprises oligonucleotide primers, dual-labeled hydrolysis TaqMan® probes and non-infectious positive control material specially designed for the specific detection and assessment of 2019-nCoV viral RNA in respiratory specimens.
The two sets of oligonucleotide primers and labeled TaqMan® probes (2019-nCoV_N1 and 2019-nCoV_N2) were selected from regions of the virus nucleocapsid (N) gene. An additional primer/probe set to detect the human RNase P gene (RP) in control samples and clinical specimens is also provided in the CDC’s laboratory test kit. TaqMan® probes are labeled at the 5'-end with the reporter molecule 6-carboxyfluorescein (FAM) and with the quencher, Black Hole Quencher 1 (BHQ-1) (Biosearch Technologies, Inc., Novato, CA) at the 3'end. Alternatively, they can also be labeled at the 5'-end with the reporter molecule 6-carboxyfluorescein (FAM) and with a double quencher, ZENTM Internal Quencher positioned between the ninth (9th) and tenth (10th) nucleotide base in the oligonucleotide sequence and Iowa Black® FQ (3IABkFQ) located at the 3’-end (Integrated DNA Technologies, Coralville, IA). The CDC 2019-nCoV Positive Control (nCoVPC) consists of in vitro transcribed RNA that will yield a positive result with each assay including RP.
The 2019-nCoV Real-Time RT-PCR Diagnostic Panel requires the use of additional materials that are not provided with the test kit. These materials comprise PCR reagents and a control material termed Human Specimen Control (HSC). The HSC, manufactured by CDC, consists of non-infectious human cell culture preparation used as a nucleic acid extraction procedural control to demonstrate successful recovery of nucleic acid as well as extraction reagent integrity. RNA from HSC must be extracted and processed with each specimen extraction run. Regarding PCR reagents, the designed workflow uses ready-to-use master mixes for probe-based RT-qPCR from Promega.
Promega GoTaq® Probe 1- Step RT-qPCR System
The GoTaq® Probe 1-Step RT-qPCR System (Figure 1) provides optimal conditions to perform quantitative PCR assays in the hydrolysis probe detection format. The system enables detection and relative quantification of RNA expression levels using a one-step RT-qPCR method, combining GoScript™ Reverse Transcriptase and GoTaq® Probe qPCR Master Mix in single-step real-time amplification reactions. The GoTaq® Probe qPCR Master Mix contains all components for qPCR, including GoTaq® Hot Start Polymerase.
To monitor for reagent and PCR set-up contamination, a No Template Control, typically referred to as an NTC, is prepared for each run. NTC includes all PCR reagents except the DNA template which is replaced by an equivalent volume of nuclease-free water.
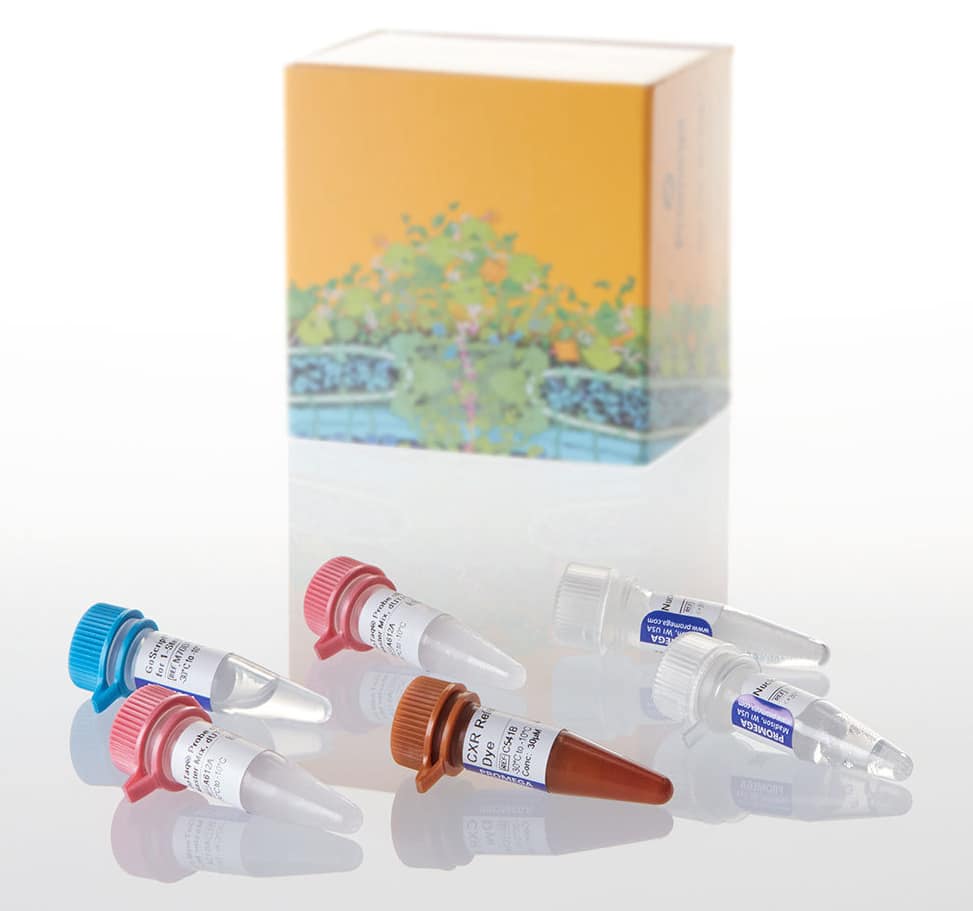
Figure 1: GoTaq® Probe 1-Step RT-qPCR System kit from Promega Corporation enabling reverse transcription and qPCR in one reaction. (Cat.# A6121)
Experimental Procedure
Total RNA isolated and purified from upper and lower respiratory specimens is reverse transcribed to cDNA by GoScript™ Reverse Transcriptase and subsequently amplified by GoTaq® Hot Start Polymerase using the Applied Biosystems 7500 Fast Dx Real-Time PCR Instrument.
In the process, the fluorescently-labeled (TaqMan®) probe anneals to a specific target sequence located between the forward and reverse primers (Figure 2). During the extension phase of the PCR cycle, the 5’ nuclease activity of Taq polymerase degrades the probe, causing the reporter dye to separate from the quencher dye thereby generating a fluorescent signal (Figure 2). With each cycle, additional reporter dye molecules are cleaved from their respective probes, increasing the fluorescence intensity, which is monitored in real time at each PCR cycle.
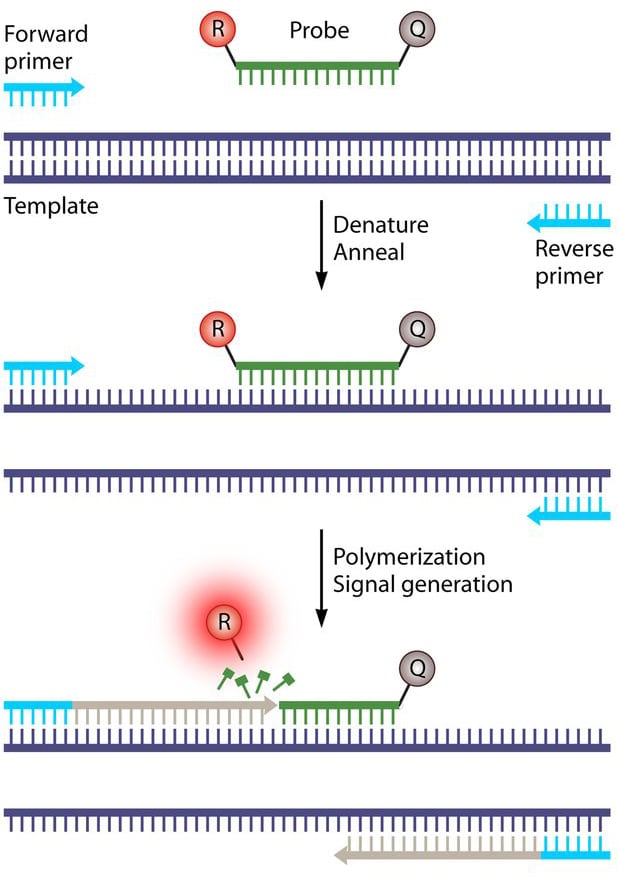
Figure 2: Hydrolysis or TaqMan probe-based real-time PCR chemistry. Image adapted from (1).
Automation of Reaction Preparation for RT-qPCR using the Andrew+ Pipetting Robot
The OneLab adaptation of The CDC 2019-nCoV Real-Time RT-PCR Diagnostic Panel can be executed manually using Pipette+ thus benefiting from the step-by-step guidance but is also amenable for automation using the Andrew+ robot. The Domino/Device+ configuration and pipetting settings were verified and validated using Andrew+. The streamlined workflow decreases hands-on time and minimizes the risk of pipetting error thereby improving overall efficiency and data quality. Moreover, OneLab software guarantees full traceability of protocol execution.
Materials Included in the Diagnostic Panel
The CDC 2019-nCoV Real-Time RT-PCR Diagnostic Panel comprises two boxes containing four reagents in total (Figure 3).
Catalog #2019-nCoVEUA-01 Diagnostic Panel Oligonucleotide Box
Three combined primer/probe sets, dried:
- 2019-nCoV_N1: target virus nucleocapsid (N) gene for specific detection of SARS-CoV-2
- 2019-nCoV_N2: target virus nucleocapsid (N) gene for specific detection of SARS-CoV-2
- RP: target human RNase P gene for detection of human nucleic acids; control for sample integrity
Catalog #2019-nCoVEUA-01 Diagnostic Panel Control Box
- Four vials of 2019-nCoV Positive Control (nCoVPC), dried: non-infectious positive control material
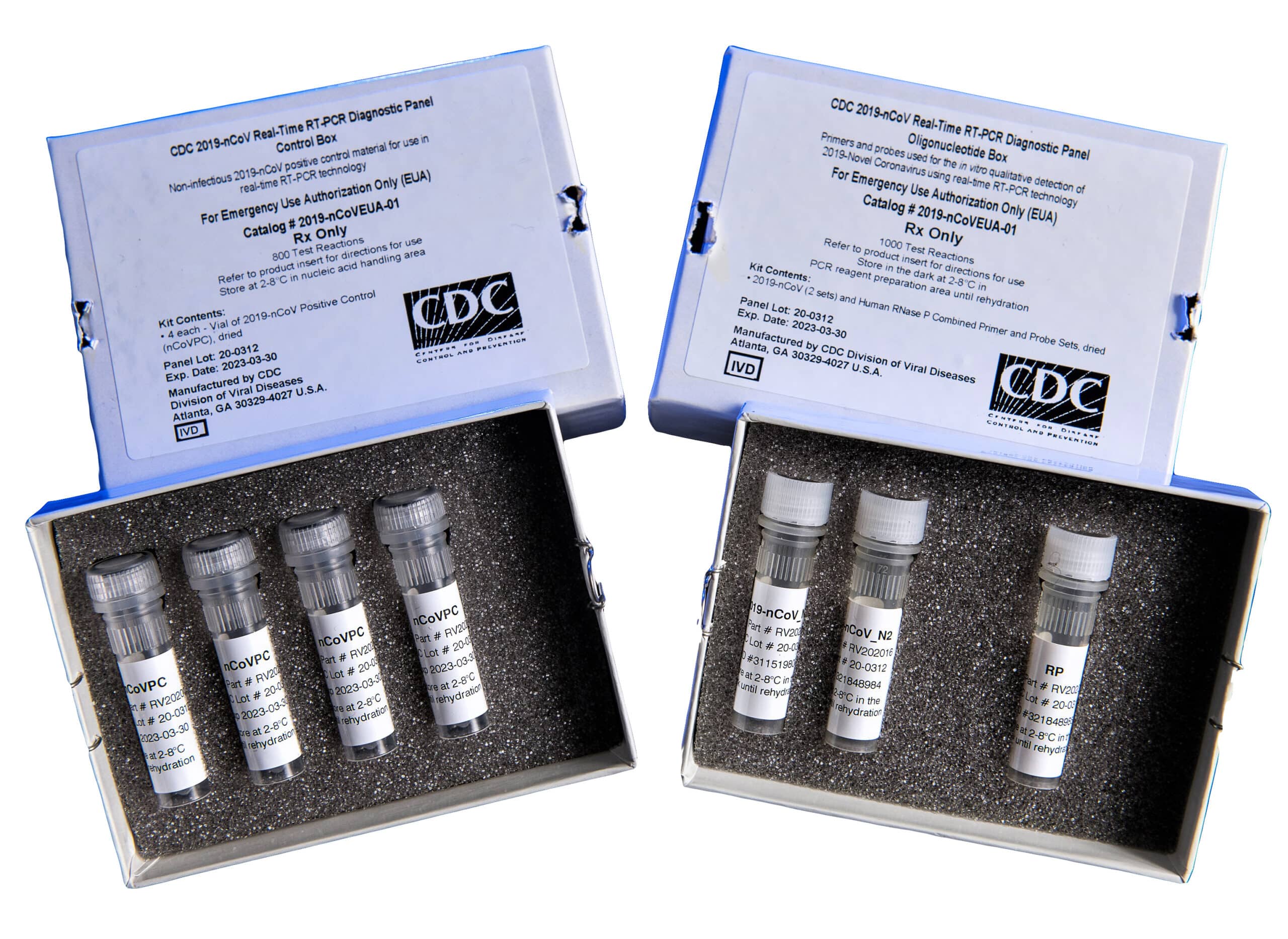
Figure 3: Boxes included in the CDC’s laboratory test kit for SARS-CoV-2.
(1) Verweij, Jaco J., and C. Rune Stensvold. 2014. ‘Molecular Testing for Clinical Diagnosis and Epidemiological Investigations of Intestinal Parasitic Infections’. Clinical Microbiology Reviews 27 (2): 371–418.
About CDC’s Diagnostic Test for COVID-19 and Supplies
CDC Diagnostic Panel's Instructions for Use (CDC-006-00019, Revision 05)
Protocols

Contact info

 This is an open access protocol distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited.
This is an open access protocol distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited. 